Widespread Effects of Chemokine 3' Untranslated Regions on mRNA Degradation and Protein Production in Human Cells
- PMID: 29907706
- PMCID: PMC6057839
- DOI: 10.4049/jimmunol.1800114
Widespread Effects of Chemokine 3' Untranslated Regions on mRNA Degradation and Protein Production in Human Cells
Abstract
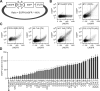
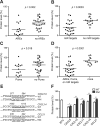
Chemokines are a large family of chemotactic cytokines that play critical roles in inflammation, development, and diseases. Chemokine expression is highly regulated during development and in response to environmental stimuli. The 3' untranslated regions (3'-UTRs) of mRNA are believed to be important in the control of chemokine gene expression. However, the regulatory effects of most chemokine 3'-UTRs have not been characterized previously. In this work, we systematically studied the effects of 43 CC and CXC chemokine 3'-UTRs on gene expression in eight human cell lines and two types of human primary cells. We found that chemokine 3'-UTRs had a wide spectrum of regulatory effects on mRNA abundance and protein production that were tightly correlated with the effects on mRNA stability. In general, 3'-UTRs had remarkably similar effects across all cell types studied. The presence of AU-rich elements, microRNA targets, and Pumilio binding sites were associated with chemokine 3'-UTR activity but did not fully account for all 3'-UTR activity detected using the reporter assay. Mutational analysis illustrated how specific cis-regulatory elements contributed to the regulatory effect of chemokine 3'-UTRs. These findings bring new insights into the mechanisms by which chemokine expression is regulated by 3'-UTRs.
Copyright © 2018 by The American Association of Immunologists, Inc.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. The New England journal of medicine. 2006;354:610–621. - PubMed
-
- Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nature immunology. 2001;2:123–128. - PubMed
-
- Gerber BO, Zanni MP, Uguccioni M, Loetscher M, Mackay CR, Pichler WJ, Yawalkar N, Baggiolini M, Moser B. Functional expression of the eotaxin receptor CCR3 in T lymphocytes co-localizing with eosinophils. Current biology : CB. 1997;7:836–843. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

