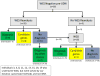
A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative
- PMID: 29907797
- PMCID: PMC6295275
- DOI: 10.1038/s41436-018-0044-2
A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative
Abstract
Purpose: Sixty to seventy-five percent of individuals with rare and undiagnosed phenotypes remain undiagnosed after exome sequencing (ES). With standard ES reanalysis resolving 10-15% of the ES negatives, further approaches are necessary to maximize diagnoses in these individuals.
Methods: In 38 ES negative patients an individualized genomic-phenotypic approach was employed utilizing (1) phenotyping; (2) reanalyses of FASTQ files, with innovative bioinformatics; (3) targeted molecular testing; (4) genome sequencing (GS); and (5) conferring of clinical diagnoses when pathognomonic clinical findings occurred.
Results: Certain and highly likely diagnoses were made in 18/38 (47%) individuals, including identifying two new developmental disorders. The majority of diagnoses (>70%) were due to our bioinformatics, phenotyping, and targeted testing identifying variants that were undetected or not prioritized on prior ES. GS diagnosed 3/18 individuals with structural variants not amenable to ES. Additionally, tentative diagnoses were made in 3 (8%), and in 5 individuals (13%) candidate genes were identified. Overall, diagnoses/potential leads were identified in 26/38 (68%).
Conclusions: Our comprehensive approach to ES negatives maximizes the ES and clinical data for both diagnoses and candidate gene identification, without GS in the majority. This iterative approach is cost-effective and is pertinent to the current conundrum of ES negatives.
Keywords: Exome sequencing; Genome sequencing; Phenotyping; Rare diseases; Undiagnosed diseases.
Conflict of interest statement
David Goldstein is a founder of and holds equity in Pairnomix and Praxis, serves as a consultant to AstraZeneca, and has research supported by Janssen, Gilead, Biogen, AstraZeneca, and UCB.
The rest of the authors declare no conflicts of interest related to this manuscript.
Figures
References
-
- Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2017;19(2):209–214. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical