Analysis of human C24 bile acids metabolome in serum and urine based on enzyme digestion of conjugated bile acids and LC-MS determination of unconjugated bile acids
- PMID: 29907951
- PMCID: PMC6424582
- DOI: 10.1007/s00216-018-1183-7
Analysis of human C24 bile acids metabolome in serum and urine based on enzyme digestion of conjugated bile acids and LC-MS determination of unconjugated bile acids
Abstract
Host-gut microbiota metabolic interactions are closely associated with health and disease. A manifestation of such co-metabolism is the vast structural diversity of bile acids (BAs) involving both oxidative stereochemistry and conjugation. Herein, we describe the development and validation of a LC-MS-based method for the analysis of human C24 BA metabolome in serum and urine. The method has high throughput covering the discrimination of oxidative stereochemistry of unconjugated species in a 15-min analytical cycle. The validated quantitative performance provided an indirect way to ascertain the conjugation patterns of BAs via enzyme-digestion protocols that incorporated the enzymes, sulfatase, β-glucuronidase, and choloylglycine hydrolase. Application of the method has led to the detection of at least 70 unconjugated BAs including 27 known species and 43 newly found species in the post-prandial serum and urine samples from 7 nonalcoholic steatohepatitis patients and 13 healthy volunteers. Newly identified unconjugated BAs included 3α, 12β-dihydroxy-5β-cholan-24-oic acid, 12α-hydroxy-3-oxo-5β-cholan-24-oic acid, and 3α, 7α, 12β-trihydroxy-5β-cholan-24-oic acid. High-definition negative fragment spectra of the other major unknown species were acquired to facilitate future identification endeavors. An extensive conjugation pattern is the major reason for the "invisibility" of the newly found BAs to other common analytical methods. Metabolomic analysis of the total unconjugated BA profile in combination with analysis of their conjugation patterns and urinary excretion tendencies have provided substantial insights into the interconnected roles of host and gut microbiota in maintaining BA homeostasis. It was proposed that the urinary total BA profile may serve as an ideal footprint for the functional status of the host-gut microbial BA co-metabolism. In summary, this work provided a powerful tool for human C24 BA metabolome analysis that bridges the gap between GC-MS techniques in the past age and LC-MS techniques currently prevailing in biomedical researches. Further applications of the present method in clinical, translational research, and other biomedical explorations will continue to boost the construction of a host-gut microbial co-metabolism network of BAs and thus facilitate the decryption of BA-mediated host-gut microbiota crosstalk in health and diseases. Graphical abstract ᅟ.
Keywords: Bile acid; Conjugation; Host-gut microbial co-metabolism; Liquid chromatography; Oxidative stereochemistry; Tandem mass spectrometry.
Conflict of interest statement
Compliance with Ethical Standards
The authors declare that there are no conflicts of interest. The human urine and serum samples used in this work was collected from the clinical trial approved by the UNC-CH Biomedical Institutional Review Board and published in
Figures
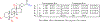

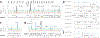
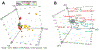

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

