Diet Modifies Colonic Microbiota and CD4+ T-Cell Repertoire to Induce Flares of Colitis in Mice With Myeloid-Cell Expression of Interleukin 23
- PMID: 29909020
- PMCID: PMC6174107
- DOI: 10.1053/j.gastro.2018.06.034
Diet Modifies Colonic Microbiota and CD4+ T-Cell Repertoire to Induce Flares of Colitis in Mice With Myeloid-Cell Expression of Interleukin 23
Abstract
Background & aims: Several studies have shown that signaling via the interleukin 23 (IL23) receptor is required for development of colitis. We studied the roles of IL23, dietary factors, alterations to the microbiota, and T cells in the development and progression of colitis in mice.
Methods: All mice were maintained on laboratory diet 5053, unless otherwise noted. We generated mice that express IL23 in CX3CR1-positive myeloid cells (R23FR mice) upon cyclic administration of tamoxifen dissolved in diet 2019. Diets 2019 and 5053 have minor differences in the overall composition of protein, fat, fiber, minerals, and vitamins. CX3CR1CreER mice (FR mice) were used as controls. Some mice were given antibiotics, and others were raised in a germ-free environment. Intestinal tissues were collected and analyzed by histology and flow cytometry. Feces were collected and analyzed by 16S rDNA sequencing. Feces from C57/Bl6, R23FR, or FR mice were fed to FR and R23FR germ-free mice in microbiota transplant experiments. We also performed studies with R23FR/Rag-/-, R23FR/Mu-/-, and R23FR/Tcrd-/- mice. R23FR mice were given injections of antibodies against CD4 or CD8 to deplete T cells. Mesenteric lymph nodes and large intestine CD4+ cells from R23FR or FR mice in remission from colitis were transferred into Rag-/- mice. CD4+ cells were isolated from donor R23FR mice and recipient Rag-/- mice, and T-cell receptor sequences were determined.
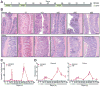
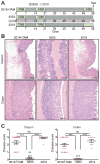
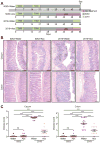
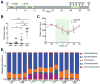
Results: Expression of IL23 led to development of a relapsing-remitting colitis that was dependent on the microbiota and CD4+ T cells. The relapses were caused by switching from the conventional diet used in our facility (diet 5053) to the diet 2019 and were not dependent on tamoxifen after the first cycle. The switch in the diet modified the microbiota but did not alter levels of IL23 in intestinal tissues compared with mice that remained on the conventional diet. Mesenteric lymph nodes and large intestine CD4+ cells from R23FR mice in remission, but not from FR mice, induced colitis after transfer into Rag-/- mice, but only when these mice were placed on the diet 2019. The CD4+ T-cell receptor repertoire of Rag-/- mice with colitis (fed the 2019 diet) was less diverse than that from donor mice and Rag-/- mice without colitis (fed the 5053 diet) because of expansion of dominant T-cell clones.
Conclusions: We developed mice that express IL23 in CX3CR1-positive myeloid cells (R23FR mice) and found that they are more susceptible to diet-induced colitis than mice that do not express IL23. The R23FR mice have a population of CD4+ T cells that becomes activated in response to dietary changes and alterations to the intestinal microbiota. The results indicate that alterations in the diet, intestinal microbiota, and IL23 signaling can contribute to pathogenesis of inflammatory bowel disease.
Keywords: Cytokine; Immune Response; Inflammatory Bowel Disease Model; Microbiome.
Copyright © 2018 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Both R.S. and A.J. are co-founders of Girihlet Inc., which has licensed the TCR sequencing technology from Mount Sinai, with the goal of developing it as a commercial product. JF.C. has served as consultant, advisory board member or speaker for AbbVie, Amgen, Boehringer-Ingelheim, Celgene Corporation, Celltrion, Enterome, Ferring, Genentech, Janssen and Janssen, Lilly, Medimmune, Merck & Co., Pfizer, PPM Services, Protagonist, Second Genome, Seres, Shire, Takeda, Theradiag, Theravance Biopharma. Stock options: Intestinal Biotech Development, Genfit. JF.C. has research Grants from AbbVie, Takeda, Janssen. The remaining authors disclose no conflicts.
Figures


References
-
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–2778. - PubMed
-
- Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nature Reviews Immunology. 2013;13:321–335. - PubMed
-
- Nishikawa J, Kudo T, Sakata S, et al. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scandinavian Journal of Gastroenterology. 2009;44:180–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

