Mitochondrial protein import regulates cytosolic protein homeostasis and neuronal integrity
- PMID: 29909722
- PMCID: PMC6208451
- DOI: 10.1080/15548627.2018.1474991
Mitochondrial protein import regulates cytosolic protein homeostasis and neuronal integrity
Abstract
Neurodegeneration is characterized by protein aggregate deposits and mitochondrial malfunction. Reduction in Tom40 (translocase of outer membrane 40) expression, a key subunit of the translocase of the outer mitochondrial membrane complex, led to accumulation of ubiquitin (Ub)-positive protein aggregates engulfed by Atg8a-positive membranes. Other macroautophagy markers were also abnormally accumulated. Autophagy was induced but the majority of autophagosomes failed to fuse with lysosomes when Tom40 was downregulated. In Tom40 RNAi tissues, autophagosome-like (AL) structures, often not sealed, were 10 times larger than starvation induced autophagosomes. Atg5 downregulation abolished Tom40 RNAi induced AL structure formation, but the Ub-positive aggregates remained, whereas knock down of Syx17, a gene required for autophagosome-lysosome fusion, led to the disappearance of giant AL structures and accumulation of small autophagosomes and phagophores near the Ub-positive aggregates. The protein aggregates contained many mitochondrial preproteins, cytosolic proteins, and proteasome subunits. Proteasome activity and ATP levels were reduced and the ROS levels was increased in Tom40 RNAi tissues. The simultaneous inhibition of proteasome activity, reduction in ATP production, and increase in ROS, but none of these conditions alone, can mimic the imbalanced proteostasis phenotypes observed in Tom40 RNAi cells. Knockdown of ref(2)P or ectopic expression of Pink1 and park greatly reduced aggregate formation in Tom40 RNAi tissues. In nerve tissues, reduction in Tom40 activity leads to aggregate formation and neurodegeneration. Rather than diminishing the neurodegenerative phenotypes, overexpression of Pink1 enhanced them. We proposed that defects in mitochondrial protein import may be the key to linking imbalanced proteostasis and mitochondrial defects.
Abbreviations: AL: autophagosome-like; Atg12: Autophagy-related 12; Atg14: Autophagy-related 14; Atg16: Autophagy-related 16; Atg5: Autophagy-related 5; Atg6: Autophagy-related 6; Atg8a: Autophagy-related 8a; Atg9: Autophagy-related 9; ATP: adenosine triphosphate; Cas9: CRISPR associated protein 9; cDNA: complementary DNA; COX4: Cytochrome c oxidase subunit 4; CRISPR: clustered regularly interspaced short palindromic repeats; Cyt-c1: Cytochrome c1; DAPI: 4,6-diamidino-2-phenylindole dihydrochloride; Dcr-2: Dicer-2; FLP: Flippase recombination enzyme; FRT: FLP recombination target; GFP: green fluorescent protein; GO: gene ontology; gRNA: guide RNA; Hsp60: Heat shock protein 60A; HDAC6: Histone deacetylase 6; htt: huntingtin; Idh: Isocitrate dehydrogenase; IFA: immunofluorescence assay; Irp-1A: Iron regulatory protein 1A; kdn: knockdown; Marf: Mitochondrial assembly regulatory factor; MitoGFP: Mitochondrial-GFP; MS: mass spectrometry; MTPAP: mitochondrial poly(A) polymerase; Nmnat: Nicotinamide mononucleotide adenylyltransferase; OE: overexpression; Pink1/PINK1: PTEN-induced putative kinase 1; polyQ: polyglutamine; PRKN: parkin RBR E3 ubiquitin protein ligase; Prosα4: proteasome α4 subunit; Prosβ1: proteasome β1 subunit; Prosβ5: proteasome β5 subunit; Prosβ7: proteasome β7 subunit; ref(2)P: refractory to sigma P; RFP: red fluorescent protein; RNAi: RNA interference; ROS: reactive oxygen species; Rpn11: Regulatory particle non-ATPase 11; Rpt2: Regulatory particle triple-A ATPase 2; scu: scully; sicily: severe impairment of CI with lengthened youth; sesB: stress-sensitive B; Syx17: Syntaxin17; TEM: transmission electron microscopy; ttm50: tiny tim 50; Tom: translocase of the outer membrane; Tom20: translocase of outer membrane 20; Tom40: translocase of outer membrane 40; Tom70: translocase of outer membrane 70; UAS: upstream active sequence; Ub: ubiquitin; VNC: ventral nerve cord; ZFYVE1: zinc finger FYVE-type containing 1.
Keywords: Autophagy; TOM Complex; drosophila; mitochondria; neurodegeneration; proteinaggregates.
Figures
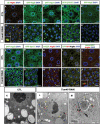

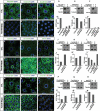
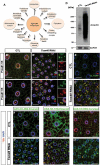

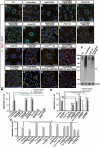

References
-
- Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr Opin Neurobiol. 2007. June;17(3):331–337. S0959-4388(07)00064-5 [pii]. PubMed PMID: 17499497; eng. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
