Phylogeny of dermatophytes with genomic character evaluation of clinically distinct Trichophyton rubrum and T. violaceum
- PMID: 29910521
- PMCID: PMC6002342
- DOI: 10.1016/j.simyco.2018.02.004
Phylogeny of dermatophytes with genomic character evaluation of clinically distinct Trichophyton rubrum and T. violaceum
Abstract
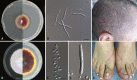
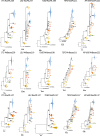
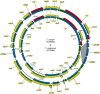
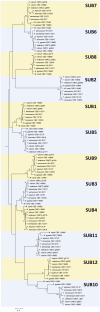
Trichophyton rubrum and T. violaceum are prevalent agents of human dermatophyte infections, the former being found on glabrous skin and nail, while the latter is confined to the scalp. The two species are phenotypically different but are highly similar phylogenetically. The taxonomy of dermatophytes is currently being reconsidered on the basis of molecular phylogeny. Molecular species definitions do not always coincide with existing concepts which are guided by ecological and clinical principles. In this article, we aim to bring phylogenetic and ecological data together in an attempt to develop new species concepts for anthropophilic dermatophytes. Focus is on the T. rubrum complex with analysis of rDNA ITS supplemented with LSU, TUB2, TEF3 and ribosomal protein L10 gene sequences. In order to explore genomic differences between T. rubrum and T. violaceum, one representative for both species was whole genome sequenced. Draft sequences were compared with currently available dermatophyte genomes. Potential virulence factors of adhesins and secreted proteases were predicted and compared phylogenetically. General phylogeny showed clear gaps between geophilic species of Arthroderma, but multilocus distances between species were often very small in the derived anthropophilic and zoophilic genus Trichophyton. Significant genome conservation between T. rubrum and T. violaceum was observed, with a high similarity at the nucleic acid level of 99.38 % identity. Trichophyton violaceum contains more paralogs than T. rubrum. About 30 adhesion genes were predicted among dermatophytes. Seventeen adhesins were common between T. rubrum and T. violaceum, while four were specific for the former and eight for the latter. Phylogenetic analysis of secreted proteases reveals considerable expansion and conservation among the analyzed species. Multilocus phylogeny and genome comparison of T. rubrum and T. violaceum underlined their close affinity. The possibility that they represent a single species exhibiting different phenotypes due to different localizations on the human body is discussed.
Keywords: Adhesion; Arthrodermataceae; Character analysis; Dermatophytes; Genome; Phylogeny; Protease; Trichophyton rubrum; Trichophyton violaceum.
Figures




Similar articles
-
Expression Profiles of Protease in Onychomycosis-Related Pathogenic Trichophyton rubrum and Tinea Capitis-Related Pathogenic Trichophyton violaceum.Mycopathologia. 2024 Jan 24;189(1):14. doi: 10.1007/s11046-024-00828-3. Mycopathologia. 2024. PMID: 38265566
-
Toward a Novel Multilocus Phylogenetic Taxonomy for the Dermatophytes.Mycopathologia. 2017 Feb;182(1-2):5-31. doi: 10.1007/s11046-016-0073-9. Epub 2016 Oct 25. Mycopathologia. 2017. PMID: 27783317 Free PMC article.
-
Differences among chitin synthase I gene sequences in Trichophyton rubrum and T. violaceum.Med Mycol. 2000 Feb;38(1):47-50. doi: 10.1080/mmy.38.1.47.50. Med Mycol. 2000. PMID: 10746227
-
Mycology - an update. Part 1: Dermatomycoses: causative agents, epidemiology and pathogenesis.J Dtsch Dermatol Ges. 2014 Mar;12(3):188-209; quiz 210, 188-211; quiz 212. doi: 10.1111/ddg.12245. Epub 2014 Feb 17. J Dtsch Dermatol Ges. 2014. PMID: 24533779 Review.
-
[Trichophyton violaceum : Main cause of tinea capitis in children at Mbarara Regional Referral Hospital in Uganda].Hautarzt. 2016 Sep;67(9):712-7. doi: 10.1007/s00105-016-3831-1. Hautarzt. 2016. PMID: 27341825 Review. German.
Cited by
-
A Polyphasic Approach to Classification and Identification of Species within the Trichophyton benhamiae Complex.J Fungi (Basel). 2021 Jul 26;7(8):602. doi: 10.3390/jof7080602. J Fungi (Basel). 2021. PMID: 34436141 Free PMC article.
-
A Comparative Description of Dermatophyte Genomes: A State-of-the-Art Review.Mycopathologia. 2023 Dec;188(6):1007-1025. doi: 10.1007/s11046-023-00802-5. Epub 2023 Oct 9. Mycopathologia. 2023. PMID: 37812320
-
Identification of clinical dermatophyte isolates obtained from Iran by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.Curr Med Mycol. 2019 Jun;5(2):22-26. doi: 10.18502/cmm.5.2.1157. Curr Med Mycol. 2019. PMID: 31321334 Free PMC article.
-
Updating the Taxonomy of Dermatophytes of the BCCM/IHEM Collection According to the New Standard: A Phylogenetic Approach.Mycopathologia. 2020 Feb;185(1):161-168. doi: 10.1007/s11046-019-00338-7. Epub 2019 May 15. Mycopathologia. 2020. PMID: 31093849
-
Molecular Epidemiology, Genetic Diversity, and Antifungal Susceptibility of Major Pathogenic Dermatophytes Isolated From Human Dermatophytosis.Front Microbiol. 2021 Jun 4;12:643509. doi: 10.3389/fmicb.2021.643509. eCollection 2021. Front Microbiol. 2021. PMID: 34149634 Free PMC article.
References
-
- Ates A.N., Ozcan K., Ilkit M.T. Diagnostic value of morphological, physiological and biochemical tests in distinguishing Trichophyton rubrum from Trichophyton mentagrophytes complex. Medical Mycology. 2008;46:811–822. - PubMed
-
- Ayanbimpe G.M., Taghir H., Diya A. Tinea capitis among primary school children in some parts of central Nigeria. Mycoses. 2008;51:336–340. - PubMed
-
- Bolognia J.L., Jorizzo J.L., Rapinl R.P. 2nd ed. Elsevier; London, UK: 2008. Dermatology.
LinkOut - more resources
Full Text Sources
Other Literature Sources
