A Review of Notch Processing With New Insights Into Ligand-Independent Notch Signaling in T-Cells
- PMID: 29910816
- PMCID: PMC5992298
- DOI: 10.3389/fimmu.2018.01230
A Review of Notch Processing With New Insights Into Ligand-Independent Notch Signaling in T-Cells
Abstract
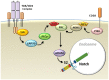
The Notch receptor is an evolutionarily highly conserved transmembrane protein essential to a wide spectrum of cellular systems, and its deregulation has been linked to a vast number of developmental disorders and malignancies. Regulated Notch function is critical for the generation of T-cells, in which abnormal Notch signaling results in leukemia. Notch activation through trans-activation of the receptor by one of its ligands expressed on adjacent cells has been well defined. In this canonical ligand-dependent pathway, Notch receptor undergoes conformational changes upon ligand engagement, stimulated by a pulling-force on the extracellular fragment of Notch that results from endocytosis of the receptor-bound ligand into the ligand-expressing cell. These conformational changes in the receptor allow for two consecutive proteolytic cleavage events to occur, which release the intracellular region of the receptor into the cytoplasm. It can then travel to the nucleus, where it induces gene transcription. However, there is accumulating evidence that other pathways may induce Notch signaling. A ligand-independent mechanism of Notch activation has been described in which receptor processing is initiated via cell-internal signals. These signals result in the internalization of Notch into endosomal compartments, where chemical changes existing in this microenvironment result in the conformational modifications required for receptor processing. This review will present mechanisms underlying both canonical ligand-dependent and non-canonical ligand-independent Notch activation pathways and discuss the latter in the context of Notch signaling in T-cells.
Keywords: Notch; T-cell; T-cell receptor; endocytosis; ligand-independent; protein kinase C.
Figures
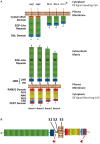
 . Abbreviations: NRR, negative regulatory region; LNR, cysteine-rich Lin12/Notch repeats; HD, heterodimerization domain; NLS, nuclear localization sequence; TAD, transcriptional activation domain; DSL, Delta/Serrate/Lag-2; Jag, jagged; DLL, delta-like.
. Abbreviations: NRR, negative regulatory region; LNR, cysteine-rich Lin12/Notch repeats; HD, heterodimerization domain; NLS, nuclear localization sequence; TAD, transcriptional activation domain; DSL, Delta/Serrate/Lag-2; Jag, jagged; DLL, delta-like.


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

