Mammalian TRIM67 Functions in Brain Development and Behavior
- PMID: 29911180
- PMCID: PMC6002264
- DOI: 10.1523/ENEURO.0186-18.2018
Mammalian TRIM67 Functions in Brain Development and Behavior
Erratum in
-
Erratum: Boyer et al., "Mammalian TRIM67 Functions in Brain Development and Behavior".eNeuro. 2019 Aug 16;6(4):ENEURO.0281-19.2019. doi: 10.1523/ENEURO.0281-19.2019. Print 2019 Jul/Aug. eNeuro. 2019. PMID: 31420404 Free PMC article. No abstract available.
Abstract
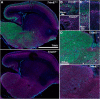
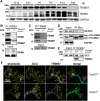
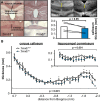
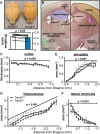
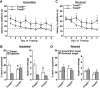
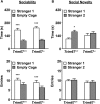
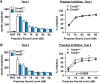
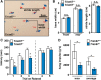
Class I members of the tripartite motif (TRIM) family of E3 ubiquitin ligases evolutionarily appeared just prior to the advent of neuronal like cells and have been implicated in neuronal development from invertebrates to mammals. The single Class I TRIM in Drosophila melanogaster and Caenorhabditis elegans and the mammalian Class I TRIM9 regulate axon branching and guidance in response to the guidance cue netrin, whereas mammalian TRIM46 establishes the axon initial segment. In humans, mutations in TRIM1 and TRIM18 are implicated in Opitz Syndrome, characterized by midline defects and often intellectual disability. We find that although TRIM67 is the least studied vertebrate Class I TRIM, it is the most evolutionarily conserved. Here we show that mammalian TRIM67 interacts with both its closest paralog TRIM9 and the netrin receptor DCC and is differentially enriched in specific brain regions during development and adulthood. We describe the anatomical and behavioral consequences of deletion of murine Trim67. While viable, mice lacking Trim67 exhibit abnormal anatomy of specific brain regions, including hypotrophy of the hippocampus, striatum, amygdala, and thalamus, and thinning of forebrain commissures. Additionally, Trim67-/- mice display impairments in spatial memory, cognitive flexibility, social novelty preference, muscle function, and sensorimotor gating, whereas several other behaviors remain intact. This study demonstrates the necessity for TRIM67 in appropriate brain development and behavior.
Keywords: DCC; TRIM67; commissure; hippocampus; knock-out; striatum.
Figures




Comment in
-
TRIMing Neural Connections with Ubiquitin.Dev Cell. 2019 Jan 7;48(1):5-6. doi: 10.1016/j.devcel.2018.12.012. Dev Cell. 2019. PMID: 30620903 Free PMC article.
References
-
- Anuppalle M, Maddirevula S, Huh TL, Rhee M (2013) Ubiquitin proteasome system networks in the neurological disorders. Animal Cells Syst (Seoul) 17:383–387. 10.1080/19768354.2013.855256 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
