Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families
- PMID: 29911924
- PMCID: PMC6546367
- DOI: 10.1080/15476286.2018.1483685
Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families
Abstract
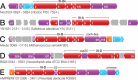
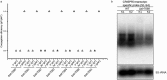
A study was undertaken to identify conserved proteins that are encoded adjacent to cas gene cassettes of Type III CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats - CRISPR associated) interference modules. Type III modules have been shown to target and degrade dsDNA, ssDNA and ssRNA and are frequently intertwined with cofunctional accessory genes, including genes encoding CRISPR-associated Rossman Fold (CARF) domains. Using a comparative genomics approach, and defining a Type III association score accounting for coevolution and specificity of flanking genes, we identified and classified 39 new Type III associated gene families. Most archaeal and bacterial Type III modules were seen to be flanked by several accessory genes, around half of which did not encode CARF domains and remain of unknown function. Northern blotting and interference assays in Synechocystis confirmed that one particular non-CARF accessory protein family was involved in crRNA maturation. Non-CARF accessory genes were generally diverse, encoding nuclease, helicase, protease, ATPase, transporter and transmembrane domains with some encoding no known domains. We infer that additional families of non-CARF accessory proteins remain to be found. The method employed is scalable for potential application to metagenomic data once automated pipelines for annotation of CRISPR-Cas systems have been developed. All accessory genes found in this study are presented online in a readily accessible and searchable format for researchers to audit their model organism of choice: http://accessory.crispr.dk .
Keywords: CARF; CRISPR; accessory; ancillary; archaea; auxillary; bacteria; csx1; csx3; helicase; nuclease; protease; type III.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
