Comparison of Strategies to Overcome Drug Resistance: Learning from Various Kingdoms
- PMID: 29912169
- PMCID: PMC6100412
- DOI: 10.3390/molecules23061476
Comparison of Strategies to Overcome Drug Resistance: Learning from Various Kingdoms
Abstract
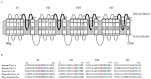
Drug resistance, especially antibiotic resistance, is a growing threat to human health. To overcome this problem, it is significant to know precisely the mechanisms of drug resistance and/or self-resistance in various kingdoms, from bacteria through plants to animals, once more. This review compares the molecular mechanisms of the resistance against phycotoxins, toxins from marine and terrestrial animals, plants and fungi, and antibiotics. The results reveal that each kingdom possesses the characteristic features. The main mechanisms in each kingdom are transporters/efflux pumps in phycotoxins, mutation and modification of targets and sequestration in marine and terrestrial animal toxins, ABC transporters and sequestration in plant toxins, transporters in fungal toxins, and various or mixed mechanisms in antibiotics. Antibiotic producers in particular make tremendous efforts for avoiding suicide, and are more flexible and adaptable to the changes of environments. With these features in mind, potential alternative strategies to overcome these resistance problems are discussed. This paper will provide clues for solving the issues of drug resistance.
Keywords: antibiotic resistance; bacterium; drug resistance; fungus; marine animal; phycotoxin; plant; self-resistance; terrestrial animal.
Conflict of interest statement
The author declares no conflict of interest.
Figures



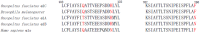

Similar articles
-
Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity.Adv Genet. 2014;85:201-53. doi: 10.1016/B978-0-12-800271-1.00004-4. Adv Genet. 2014. PMID: 24880736 Review.
-
An Atypical ABC Transporter Is Involved in Antifungal Resistance and Host Interactions in the Pathogenic Fungus Cryptococcus neoformans.mBio. 2022 Aug 30;13(4):e0153922. doi: 10.1128/mbio.01539-22. Epub 2022 Jun 21. mBio. 2022. PMID: 35726920 Free PMC article.
-
Multidrug ABC transporters in bacteria.Res Microbiol. 2019 Nov-Dec;170(8):381-391. doi: 10.1016/j.resmic.2019.06.001. Epub 2019 Jun 25. Res Microbiol. 2019. PMID: 31251973 Review.
-
Fungal transporters involved in efflux of natural toxic compounds and fungicides.Fungal Genet Biol. 2000 Jun;30(1):1-15. doi: 10.1006/fgbi.2000.1206. Fungal Genet Biol. 2000. PMID: 10955904 Review.
-
A Xenobiotic Detoxification Pathway through Transcriptional Regulation in Filamentous Fungi.mBio. 2018 Jul 17;9(4):e00457-18. doi: 10.1128/mBio.00457-18. mBio. 2018. PMID: 30018104 Free PMC article.
Cited by
-
Expression of the Stem Cell Marker ABCB5 in Normal and Tumor Tissues.In Vivo. 2022 Jul-Aug;36(4):1651-1666. doi: 10.21873/invivo.12877. In Vivo. 2022. PMID: 35738589 Free PMC article.
-
A rare urinary tract infection of multidrug-resistant Chryseobacterium urinae sp. nov. isolated from a diabetic, non-catheterized patient.Arch Microbiol. 2024 Mar 11;206(4):150. doi: 10.1007/s00203-024-03881-0. Arch Microbiol. 2024. PMID: 38466448
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

