Biochemical and Functional Characterization of Mouse Mammary Tumor Virus Full-Length Pr77Gag Expressed in Prokaryotic and Eukaryotic Cells
- PMID: 29912170
- PMCID: PMC6024702
- DOI: 10.3390/v10060334
Biochemical and Functional Characterization of Mouse Mammary Tumor Virus Full-Length Pr77Gag Expressed in Prokaryotic and Eukaryotic Cells
Abstract
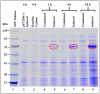
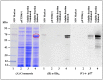
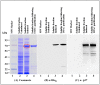
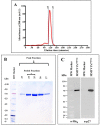
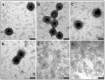
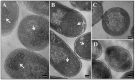
The mouse mammary tumor virus (MMTV) Pr77Gag polypeptide is an essential retroviral structural protein without which infectious viral particles cannot be formed. This process requires specific recognition and packaging of dimerized genomic RNA (gRNA) by Gag during virus assembly. Most of the previous work on retroviral assembly has used either the nucleocapsid portion of Gag, or other truncated Gag derivatives—not the natural substrate for virus assembly. In order to understand the molecular mechanism of MMTV gRNA packaging process, we expressed and purified full-length recombinant Pr77Gag-His₆-tag fusion protein from soluble fractions of bacterial cultures. We show that the purified Pr77Gag-His₆-tag protein retained the ability to assemble virus-like particles (VLPs) in vitro with morphologically similar immature intracellular particles. The recombinant proteins (with and without His₆-tag) could both be expressed in prokaryotic and eukaryotic cells and had the ability to form VLPs in vivo. Most importantly, the recombinant Pr77Gag-His₆-tag fusion proteins capable of making VLPs in eukaryotic cells were competent for packaging sub-genomic MMTV RNAs. The successful expression and purification of a biologically active, full-length MMTV Pr77Gag should lay down the foundation towards performing RNA–protein interaction(s), especially for structure-function studies and towards understanding molecular intricacies during MMTV gRNA packaging and assembly processes.
Keywords: Pr77Gag; RNA packaging; RNA–Gag interactions; RNA–protein interaction; mouse mammary tumor virus (MMTV); protein assembly; protein expression; protein purification; retrovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag.Nucleic Acids Res. 2021 May 7;49(8):4668-4688. doi: 10.1093/nar/gkab223. Nucleic Acids Res. 2021. PMID: 33836091 Free PMC article.
-
Purification and Functional Characterization of a Biologically Active Full-Length Feline Immunodeficiency Virus (FIV) Pr50Gag.Viruses. 2019 Jul 27;11(8):689. doi: 10.3390/v11080689. Viruses. 2019. PMID: 31357656 Free PMC article.
-
Expression, purification, and characterization of biologically active full-length Mason-Pfizer monkey virus (MPMV) Pr78Gag.Sci Rep. 2018 Aug 7;8(1):11793. doi: 10.1038/s41598-018-30142-0. Sci Rep. 2018. PMID: 30087395 Free PMC article.
-
Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly.Semin Cell Dev Biol. 2019 Feb;86:129-139. doi: 10.1016/j.semcdb.2018.03.015. Epub 2018 Apr 1. Semin Cell Dev Biol. 2019. PMID: 29580971 Free PMC article. Review.
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
Cited by
-
Expression, purification, and functional characterization of soluble recombinant full-length simian immunodeficiency virus (SIV) Pr55Gag.Heliyon. 2023 Jan 10;9(1):e12892. doi: 10.1016/j.heliyon.2023.e12892. eCollection 2023 Jan. Heliyon. 2023. PMID: 36685375 Free PMC article.
-
A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag.Nucleic Acids Res. 2021 May 7;49(8):4668-4688. doi: 10.1093/nar/gkab223. Nucleic Acids Res. 2021. PMID: 33836091 Free PMC article.
-
Purification and Functional Characterization of a Biologically Active Full-Length Feline Immunodeficiency Virus (FIV) Pr50Gag.Viruses. 2019 Jul 27;11(8):689. doi: 10.3390/v11080689. Viruses. 2019. PMID: 31357656 Free PMC article.
-
Role of Purine-Rich Regions in Mason-Pfizer Monkey Virus (MPMV) Genomic RNA Packaging and Propagation.Front Microbiol. 2020 Nov 5;11:595410. doi: 10.3389/fmicb.2020.595410. eCollection 2020. Front Microbiol. 2020. PMID: 33250884 Free PMC article.
-
Identification of a putative Gag binding site critical for feline immunodeficiency virus genomic RNA packaging.RNA. 2023 Dec 18;30(1):68-88. doi: 10.1261/rna.079840.123. RNA. 2023. PMID: 37914398 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

