Beyond the photocycle-how cryptochromes regulate photoresponses in plants?
- PMID: 29913346
- PMCID: PMC6240499
- DOI: 10.1016/j.pbi.2018.05.014
Beyond the photocycle-how cryptochromes regulate photoresponses in plants?
Abstract
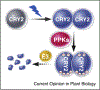
Cryptochromes (CRYs) are blue light receptors that mediate light regulation of plant growth and development. Land plants possess various numbers of cryptochromes, CRY1 and CRY2, which serve overlapping and partially redundant functions in different plant species. Cryptochromes exist as physiologically inactive monomers in darkness; photoexcited cryptochromes undergo homodimerization to increase their affinity to the CRY-signaling proteins, such as CIBs (CRY2-interacting bHLH), PIFs (Phytochrome-Interacting Factors), AUX/IAA (Auxin/INDOLE-3-ACETIC ACID), and the COP1-SPAs (Constitutive Photomorphogenesis 1-Suppressors of Phytochrome A) complexes. These light-dependent protein-protein interactions alter the activity of the CRY-signaling proteins to change gene expression and developmental programs in response to light. In the meantime, photoexcitation also changes the affinity of cryptochromes to the CRY-regulatory proteins, such as BICs (Blue-light Inhibitors of CRYs) and PPKs (Photoregulatory Protein Kinases), to modulate the activity, modification, or abundance of cryptochromes and photosensitivity of plants in response to the changing light environment.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Photoexcited CRYPTOCHROME1 Interacts with Dephosphorylated BES1 to Regulate Brassinosteroid Signaling and Photomorphogenesis in Arabidopsis.Plant Cell. 2018 Sep;30(9):1989-2005. doi: 10.1105/tpc.17.00994. Epub 2018 Aug 21. Plant Cell. 2018. PMID: 30131420 Free PMC article.
-
The dual-action mechanism of Arabidopsis cryptochromes.J Integr Plant Biol. 2024 May;66(5):883-896. doi: 10.1111/jipb.13578. Epub 2024 Jan 2. J Integr Plant Biol. 2024. PMID: 37902426 Review.
-
Mechanisms of Cryptochrome-Mediated Photoresponses in Plants.Annu Rev Plant Biol. 2020 Apr 29;71:103-129. doi: 10.1146/annurev-arplant-050718-100300. Epub 2020 Mar 13. Annu Rev Plant Biol. 2020. PMID: 32169020 Free PMC article.
-
Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana.J Plant Res. 2016 Mar;129(2):137-48. doi: 10.1007/s10265-015-0782-z. Epub 2016 Jan 25. J Plant Res. 2016. PMID: 26810763 Free PMC article. Review.
-
A CRY-BIC negative-feedback circuitry regulating blue light sensitivity of Arabidopsis.Plant J. 2017 Nov;92(3):426-436. doi: 10.1111/tpj.13664. Epub 2017 Oct 9. Plant J. 2017. PMID: 28833729 Free PMC article.
Cited by
-
Light quality as a driver of photosynthetic apparatus development.Biophys Rev. 2022 Jul 26;14(4):779-803. doi: 10.1007/s12551-022-00985-z. eCollection 2022 Aug. Biophys Rev. 2022. PMID: 36124269 Free PMC article. Review.
-
Transcriptome, metabolome and suppressor analysis reveal an essential role for the ubiquitin-proteasome system in seedling chloroplast development.BMC Plant Biol. 2022 Apr 8;22(1):183. doi: 10.1186/s12870-022-03536-6. BMC Plant Biol. 2022. PMID: 35395773 Free PMC article.
-
Shades of green: untying the knots of green photoperception.J Exp Bot. 2020 Oct 7;71(19):5764-5770. doi: 10.1093/jxb/eraa312. J Exp Bot. 2020. PMID: 32619226 Free PMC article.
-
phyB and HY5 are Involved in the Blue Light-Mediated Alleviation of Dormancy of Arabidopsis Seeds Possibly via the Modulation of Expression of Genes Related to Light, GA, and ABA.Int J Mol Sci. 2019 Nov 23;20(23):5882. doi: 10.3390/ijms20235882. Int J Mol Sci. 2019. PMID: 31771191 Free PMC article.
-
Out of the Dark and Into the Light: A New View of Phytochrome Photobodies.Front Plant Sci. 2021 Aug 31;12:732947. doi: 10.3389/fpls.2021.732947. eCollection 2021. Front Plant Sci. 2021. PMID: 34531891 Free PMC article. Review.
References
-
- Cashmore AR: Cryptochromes: enabling plants and animals to determine circadian time. Cell 2003, 114:537–543. - PubMed
-
- Sancar A: Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 2003, 103:2203–2237. - PubMed
-
- Ahmad M, Cashmore AR: HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366:162–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

