Scorpins in the DNA Damage Response
- PMID: 29914204
- PMCID: PMC6032341
- DOI: 10.3390/ijms19061794
Scorpins in the DNA Damage Response
Abstract
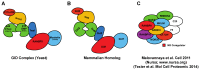
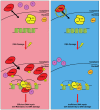
The DNA Damage Response (DDR) is a complex signaling network that comes into play when cells experience genotoxic stress. Upon DNA damage, cellular signaling pathways are rewired to slow down cell cycle progression and allow recovery. However, when the damage is beyond repair, cells activate complex and still not fully understood mechanisms, leading to a complete proliferative arrest or cell death. Several conventional and novel anti-neoplastic treatments rely on causing DNA damage or on the inhibition of the DDR in cancer cells. However, the identification of molecular determinants directing cancer cells toward recovery or death upon DNA damage is still far from complete, and it is object of intense investigation. SPRY-containing RAN binding Proteins (Scorpins) RANBP9 and RANBP10 are evolutionarily conserved and ubiquitously expressed proteins whose biological functions are still debated. RANBP9 has been previously implicated in cell proliferation, survival, apoptosis and migration. Recent studies also showed that RANBP9 is involved in the Ataxia Telangiectasia Mutated (ATM) signaling upon DNA damage. Accordingly, cells lacking RANBP9 show increased sensitivity to genotoxic treatment. Although there is no published evidence, extensive protein similarities suggest that RANBP10 might have partially overlapping functions with RANBP9. Like RANBP9, RANBP10 bears sites putative target of PIK-kinases and high throughput studies found RANBP10 to be phosphorylated following genotoxic stress. Therefore, this second Scorpin might be another overlooked player of the DDR alone or in combination with RANBP9. This review focuses on the relatively unknown role played by RANBP9 and RANBP10 in responding to genotoxic stress.
Keywords: CTLH complex; DDR; GID complex; RANBP10; RANBP9; RANBPM; Scorpins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Helena J.M., Joubert A.M., Grobbelaar S., Nolte E.M., Nel M., Pepper M.S., Coetzee M., Mercier A.E. Deoxyribonucleic Acid Damage and Repair: Capitalizing on Our Understanding of the Mechanisms of Maintaining Genomic Integrity for Therapeutic Purposes. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19041148. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

