SamSelect: a sample sequence selection algorithm for quorum planted motif search on large DNA datasets
- PMID: 29914360
- PMCID: PMC6006848
- DOI: 10.1186/s12859-018-2242-y
SamSelect: a sample sequence selection algorithm for quorum planted motif search on large DNA datasets
Abstract
Background: Given a set of t n-length DNA sequences, q satisfying 0 < q ≤ 1, and l and d satisfying 0 ≤ d < l < n, the quorum planted motif search (qPMS) finds l-length strings that occur in at least qt input sequences with up to d mismatches and is mainly used to locate transcription factor binding sites in DNA sequences. Existing qPMS algorithms have been able to efficiently process small standard datasets (e.g., t = 20 and n = 600), but they are too time consuming to process large DNA datasets, such as ChIP-seq datasets that contain thousands of sequences or more.
Results: We analyze the effects of t and q on the time performance of qPMS algorithms and find that a large t or a small q causes a longer computation time. Based on this information, we improve the time performance of existing qPMS algorithms by selecting a sample sequence set D' with a small t and a large q from the large input dataset D and then executing qPMS algorithms on D'. A sample sequence selection algorithm named SamSelect is proposed. The experimental results on both simulated and real data show (1) that SamSelect can select D' efficiently and (2) that the qPMS algorithms executed on D' can find implanted or real motifs in a significantly shorter time than when executed on D.
Conclusions: We improve the ability of existing qPMS algorithms to process large DNA datasets from the perspective of selecting high-quality sample sequence sets so that the qPMS algorithms can find motifs in a short time in the selected sample sequence set D', rather than take an unfeasibly long time to search the original sequence set D. Our motif discovery method is an approximate algorithm.
Keywords: Quorum planted motif search; Sample sequences; Transcription factor binding sites.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
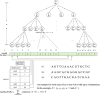
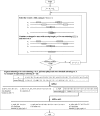


Similar articles
-
An Efficient Exact Algorithm for Planted Motif Search on Large DNA Sequence Datasets.IEEE/ACM Trans Comput Biol Bioinform. 2024 Sep-Oct;21(5):1542-1551. doi: 10.1109/TCBB.2024.3404136. Epub 2024 Oct 9. IEEE/ACM Trans Comput Biol Bioinform. 2024. PMID: 38801693
-
Improved Exact Enumerative Algorithms for the Planted (l, d)-Motif Search Problem.IEEE/ACM Trans Comput Biol Bioinform. 2014 Mar-Apr;11(2):361-74. doi: 10.1109/TCBB.2014.2306842. IEEE/ACM Trans Comput Biol Bioinform. 2014. PMID: 26355783
-
An Efficient Algorithm for Discovering Motifs in Large DNA Data Sets.IEEE Trans Nanobioscience. 2015 Jul;14(5):535-44. doi: 10.1109/TNB.2015.2421340. Epub 2015 Apr 9. IEEE Trans Nanobioscience. 2015. PMID: 25872217
-
An extension and novel solution to the (l,d)-motif challenge problem.Genome Inform. 2004;15(2):63-71. Genome Inform. 2004. PMID: 15706492 Review.
-
Discovering sequence motifs.Methods Mol Biol. 2008;452:231-51. doi: 10.1007/978-1-60327-159-2_12. Methods Mol Biol. 2008. PMID: 18566768 Review.
Cited by
-
Expanding the DNA-encoded library toolbox: identifying small molecules targeting RNA.Nucleic Acids Res. 2022 Jul 8;50(12):e67. doi: 10.1093/nar/gkac173. Nucleic Acids Res. 2022. PMID: 35288754 Free PMC article.
-
A Clustering Approach for Motif Discovery in ChIP-Seq Dataset.Entropy (Basel). 2019 Aug 16;21(8):802. doi: 10.3390/e21080802. Entropy (Basel). 2019. PMID: 33267515 Free PMC article.
References
-
- Weirauch MT, Yang A, Albu M, Cote A, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey M, Huang E, Mukherjee T, Chen X, Reece-Hoyes JS, Govindarajan S, Shaulsky G, Walhout AJM, Bouget FY, Ratsch G, Larrondo LF, Ecker JR, Hughes TR. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–1443. doi: 10.1016/j.cell.2014.08.009. - DOI - PMC - PubMed
-
- Pevzner PA, Sze SH. Combinatorial approaches to finding subtle signals in DNA sequences. In: Altman R, Bailey TL, editors. Proceedings of the Eighth International Conference on Intelligent Systems for Molecular Biology. California: AAAI Press; 2000. pp. 269–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

