Oxidative stress-induced chromosome breaks within the ABL gene: a model for chromosome rearrangement in nasopharyngeal carcinoma
- PMID: 29914565
- PMCID: PMC6006577
- DOI: 10.1186/s40246-018-0160-8
Oxidative stress-induced chromosome breaks within the ABL gene: a model for chromosome rearrangement in nasopharyngeal carcinoma
Abstract
Background: The mechanism underlying chromosome rearrangement in nasopharyngeal carcinoma (NPC) remains elusive. It is known that most of the aetiological factors of NPC trigger oxidative stress. Oxidative stress is a potent apoptotic inducer. During apoptosis, chromatin cleavage and DNA fragmentation occur. However, cells may undergo DNA repair and survive apoptosis. Non-homologous end joining (NHEJ) pathway has been known as the primary DNA repair system in human cells. The NHEJ process may repair DNA ends without any homology, although region of microhomology (a few nucleotides) is usually utilised by this DNA repair system. Cells that evade apoptosis via erroneous DNA repair may carry chromosomal aberration. Apoptotic nuclease was found to be associated with nuclear matrix during apoptosis. Matrix association region/scaffold attachment region (MAR/SAR) is the binding site of the chromosomal DNA loop structure to the nuclear matrix. When apoptotic nuclease is associated with nuclear matrix during apoptosis, it potentially cleaves at MAR/SAR. Cells that survive apoptosis via compromised DNA repair may carry chromosome rearrangement contributing to NPC tumourigenesis. The Abelson murine leukaemia (ABL) gene at 9q34 was targeted in this study as 9q34 is a common region of loss in NPC. This study aimed to identify the chromosome breakages and/or rearrangements in the ABL gene in cells undergoing oxidative stress-induced apoptosis.
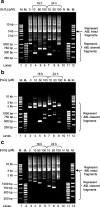
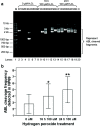
Results: In the present study, in silico prediction of MAR/SAR was performed in the ABL gene. More than 80% of the predicted MAR/SAR sites are closely associated with previously reported patient breakpoint cluster regions (BCR). By using inverse polymerase chain reaction (IPCR), we demonstrated that hydrogen peroxide (H2O2)-induced apoptosis in normal nasopharyngeal epithelial and NPC cells led to chromosomal breakages within the ABL BCR that contains a MAR/SAR. Intriguingly, we detected two translocations in H2O2-treated cells. Region of microhomology was found at the translocation junctions. This observation is consistent with the operation of microhomology-mediated NHEJ.
Conclusions: Our findings suggested that oxidative stress-induced apoptosis may participate in chromosome rearrangements of NPC. A revised model for oxidative stress-induced apoptosis mediating chromosome rearrangement in NPC is proposed.
Keywords: ABL; Apoptosis; H2O2; MAR/SAR; NPC; Oxidative stress.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures








References
-
- Shanmugaratnam K. Histological typing of nasopharyngeal carcinoma. IARC Sci Publ. 1978;20:3–12. - PubMed
-
- Hirayama T. Descriptive and analytical epidemiology of nasopharyngeal cancer. IARC Sci Publ. 1978;20:167–89. - PubMed
-
- Huang DP, Lo KW. Aetiological factors and pathogenesis. In: van Hasselt CA, Gibb AG, editors. Nasopharyngeal carcinoma. Hong Kong: The Chinese University Press; 1999. pp. 31–60.
-
- Devi BC, Pisani P, Tang TS, Parkin DM. High incidence of nasopharyngeal carcinoma in native people of Sarawak, Borneo Island. Cancer Epidemiol Biomarkers Prev. 2004;13:482–486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

