Determining the Pathogenicity of a Genomic Variant of Uncertain Significance Using CRISPR/Cas9 and Human-Induced Pluripotent Stem Cells
- PMID: 29914921
- PMCID: PMC6298866
- DOI: 10.1161/CIRCULATIONAHA.117.032273
Determining the Pathogenicity of a Genomic Variant of Uncertain Significance Using CRISPR/Cas9 and Human-Induced Pluripotent Stem Cells
Abstract
Background: The progression toward low-cost and rapid next-generation sequencing has uncovered a multitude of variants of uncertain significance (VUS) in both patients and asymptomatic "healthy" individuals. A VUS is a rare or novel variant for which disease pathogenicity has not been conclusively demonstrated or excluded, and thus cannot be definitively annotated. VUS, therefore, pose critical clinical interpretation and risk-assessment challenges, and new methods are urgently needed to better characterize their pathogenicity.
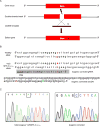
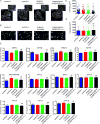
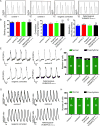
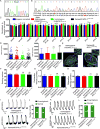
Methods: To address this challenge and showcase the uncertainty surrounding genomic variant interpretation, we recruited a "healthy" asymptomatic individual, lacking cardiac-disease clinical history, carrying a hypertrophic cardiomyopathy (HCM)-associated genetic variant (NM_000258.2:c.170C>A, NP_000249.1:p.Ala57Asp) in the sarcomeric gene MYL3, reported by the ClinVar database to be "likely pathogenic." Human-induced pluripotent stem cells (iPSCs) were derived from the heterozygous VUS MYL3(170C>A) carrier, and their genome was edited using CRISPR/Cas9 to generate 4 isogenic iPSC lines: (1) corrected "healthy" control; (2) homozygous VUS MYL3(170C>A); (3) heterozygous frameshift mutation MYL3(170C>A/fs); and (4) known heterozygous MYL3 pathogenic mutation (NM_000258.2:c.170C>G), at the same nucleotide position as VUS MYL3(170C>A), lines. Extensive assays including measurements of gene expression, sarcomere structure, cell size, contractility, action potentials, and calcium handling were performed on the isogenic iPSC-derived cardiomyocytes (iPSC-CMs).
Results: The heterozygous VUS MYL3(170C>A)-iPSC-CMs did not show an HCM phenotype at the gene expression, morphology, or functional levels. Furthermore, genome-edited homozygous VUS MYL3(170C>A)- and frameshift mutation MYL3(170C>A/fs)-iPSC-CMs lines were also asymptomatic, supporting a benign assessment for this particular MYL3 variant. Further assessment of the pathogenic nature of a genome-edited isogenic line carrying a known pathogenic MYL3 mutation, MYL3(170C>G), and a carrier-specific iPSC-CMs line, carrying a MYBPC3(961G>A) HCM variant, demonstrated the ability of this combined platform to provide both pathogenic and benign assessments.
Conclusions: Our study illustrates the ability of clustered regularly interspaced short palindromic repeats/Cas9 genome-editing of carrier-specific iPSCs to elucidate both benign and pathogenic HCM functional phenotypes in a carrier-specific manner in a dish. As such, this platform represents a promising VUS risk-assessment tool that can be used for assessing HCM-associated VUS specifically, and VUS in general, and thus significantly contribute to the arsenal of precision medicine tools available in this emerging field.
Keywords: CRISPR-Cas systems; cardiomyopathy, hypertrophic; clustered regularly interspaced short palindromic repeats; gene editing; induced pluripotent stem cells; mutations.
Conflict of interest statement
None
Figures

Similar articles
-
Studying Pathogenetic Contribution of a Variant of Unknown Significance, p.M659I (c.1977G > A) in MYH7, to the Development of Hypertrophic Cardiomyopathy Using CRISPR/Cas9-Engineered Isogenic Induced Pluripotent Stem Cells.Int J Mol Sci. 2024 Aug 9;25(16):8695. doi: 10.3390/ijms25168695. Int J Mol Sci. 2024. PMID: 39201382 Free PMC article.
-
Generation of Isogenic iPSC Lines for Studying the Effect of the p.N515del (c.1543_1545delAAC) Variant on MYBPC3 Function and Hypertrophic Cardiomyopathy Pathogenesis.Int J Mol Sci. 2024 Nov 30;25(23):12900. doi: 10.3390/ijms252312900. Int J Mol Sci. 2024. PMID: 39684611 Free PMC article.
-
iPSC-Based Modeling of Variable Clinical Presentation in Hypertrophic Cardiomyopathy.Circ Res. 2023 Jul 7;133(2):108-119. doi: 10.1161/CIRCRESAHA.122.321951. Epub 2023 Jun 15. Circ Res. 2023. PMID: 37317833
-
Deciphering pathogenicity of variants of uncertain significance with CRISPR-edited iPSCs.Trends Genet. 2021 Dec;37(12):1109-1123. doi: 10.1016/j.tig.2021.08.009. Epub 2021 Sep 8. Trends Genet. 2021. PMID: 34509299 Free PMC article. Review.
-
CRISPR/Cas9 Genome Editing: A Promising Tool for Therapeutic Applications of Induced Pluripotent Stem Cells.Curr Stem Cell Res Ther. 2018;13(4):243-251. doi: 10.2174/1574888X13666180214124800. Curr Stem Cell Res Ther. 2018. PMID: 29446747 Review.
Cited by
-
Patient-specific induced pluripotent stem cells as "disease-in-a-dish" models for inherited cardiomyopathies and channelopathies - 15 years of research.World J Stem Cells. 2021 Apr 26;13(4):281-303. doi: 10.4252/wjsc.v13.i4.281. World J Stem Cells. 2021. PMID: 33959219 Free PMC article. Review.
-
Personalized medicine in cardio-oncology: the role of induced pluripotent stem cell.Cardiovasc Res. 2019 Apr 15;115(5):949-959. doi: 10.1093/cvr/cvz024. Cardiovasc Res. 2019. PMID: 30768178 Free PMC article. Review.
-
Patient-independent human induced pluripotent stem cell model: A new tool for rapid determination of genetic variant pathogenicity in long QT syndrome.Heart Rhythm. 2019 Nov;16(11):1686-1695. doi: 10.1016/j.hrthm.2019.04.031. Epub 2019 Apr 18. Heart Rhythm. 2019. PMID: 31004778 Free PMC article.
-
Dilated cardiomyopathy in the era of precision medicine: latest concepts and developments.Heart Fail Rev. 2022 Jul;27(4):1173-1191. doi: 10.1007/s10741-021-10139-0. Epub 2021 Jul 14. Heart Fail Rev. 2022. PMID: 34263412 Free PMC article. Review.
-
Cellular models and therapeutic perspectives in hypertrophic cardiomyopathy.Med Genet. 2021 Dec 3;33(3):235-243. doi: 10.1515/medgen-2021-2094. eCollection 2021 Sep. Med Genet. 2021. PMID: 38835701 Free PMC article.
References
-
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Comm ALQA Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. - PMC - PubMed
-
- Petrucelli N, Lazebnik N, Huelsman KM, Lazebnik RS. Clinical interpretation and recommendations for patients with a variant of uncertain significance in BRCA1 or BRCA2: A survey of genetic counseling practice. Genet Test. 2002;6:107–113. - PubMed
-
- Maron BJ, Maron MS, Semsarian C. Genetics of Hypertrophic cardiomyopathy after 20 years clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

