Extinction of Zika Virus and Usutu Virus by Lethal Mutagenesis Reveals Different Patterns of Sensitivity to Three Mutagenic Drugs
- PMID: 29914957
- PMCID: PMC6125542
- DOI: 10.1128/AAC.00380-18
Extinction of Zika Virus and Usutu Virus by Lethal Mutagenesis Reveals Different Patterns of Sensitivity to Three Mutagenic Drugs
Abstract
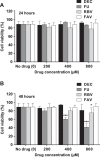
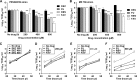
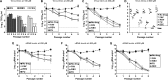
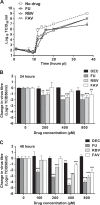
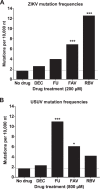
Flaviviruses constitute an increasing source of public health concern, with growing numbers of pathogens causing disease and geographic spread to temperate climates. Despite a large body of evidence supporting mutagenesis as a conceivable antiviral strategy, there are currently no data on the sensitivity to increased mutagenesis for Zika virus (ZIKV) and Usutu virus (USUV), two emerging flaviviral threats. In this study, we demonstrate that both viruses are sensitive to three ribonucleosides, favipiravir, ribavirin, and 5-fluorouracil, that have shown mutagenic activity against other RNA viruses while remaining unaffected by a mutagenic deoxyribonucleoside. Serial cell culture passages of ZIKV in the presence of these compounds resulted in the rapid extinction of infectivity, suggesting elevated sensitivity to mutagenesis. USUV extinction was achieved when a 10-fold dilution was applied between every passage, but not in experiments involving undiluted virus, indicating an overall lower susceptibility than ZIKV. Although the two viruses are inhibited by the same three drugs, ZIKV is relatively more susceptive to serial passage in the presence of purine analogues (favipiravir and ribavirin), while USUV replication is suppressed more efficiently by 5-fluorouracil. These differences in sensitivity typically correlate with the increases in the mutation frequencies observed in each nucleoside treatment. These results are relevant to the development of efficient therapies based on lethal mutagenesis and support the rational selection of different mutagenic nucleosides for each pathogen. We will discuss the implications of these results to the fidelity of flavivirus replication and the design of antiviral therapies based on lethal mutagenesis.
Keywords: 5-fluorouracil; Usutu virus; Zika virus; error threshold; favipiravir; flavivirus; lethal mutagenesis; mutation frequency; ribavirin.
Copyright © 2018 Bassi et al.
Figures
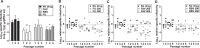

References
-
- Dash AP, Bhatia R, Sunyoto T, Mourya DT. 2013. Emerging and re-emerging arboviral diseases in Southeast Asia. J Vector Borne Dis 50:77–84. - PubMed
-
- Bogoch II, Brady OJ, Kraemer MUG, German M, Creatore MI, Brent S, Watts AG, Hay SI, Kulkarni MA, Brownstein JS, Khan K. 2016. Potential for Zika virus introduction and transmission in resource-limited countries in Africa and the Asia-Pacific region: a modelling study. Lancet Infect Dis 16:1237–1245. doi: 10.1016/S1473-3099(16)30270-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

