σI from Bacillus subtilis: Impact on Gene Expression and Characterization of σI-Dependent Transcription That Requires New Types of Promoters with Extended -35 and -10 Elements
- PMID: 29914988
- PMCID: PMC6088155
- DOI: 10.1128/JB.00251-18
σI from Bacillus subtilis: Impact on Gene Expression and Characterization of σI-Dependent Transcription That Requires New Types of Promoters with Extended -35 and -10 Elements
Abstract
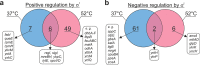
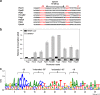
The σI sigma factor from Bacillus subtilis is a σ factor associated with RNA polymerase (RNAP) that was previously implicated in adaptation of the cell to elevated temperature. Here, we provide a comprehensive characterization of this transcriptional regulator. By transcriptome sequencing (RNA-seq) of wild-type (wt) and σI-null strains at 37°C and 52°C, we identified ∼130 genes affected by the absence of σI Further analysis revealed that the majority of these genes were affected indirectly by σI The σI regulon, i.e., the genes directly regulated by σI, consists of 16 genes, of which eight (the dhb and yku operons) are involved in iron metabolism. The involvement of σI in iron metabolism was confirmed phenotypically. Next, we set up an in vitro transcription system and defined and experimentally validated the promoter sequence logo that, in addition to -35 and -10 regions, also contains extended -35 and -10 motifs. Thus, σI-dependent promoters are relatively information rich in comparison with most other promoters. In summary, this study supplies information about the least-explored σ factor from the industrially important model organism B. subtilisIMPORTANCE In bacteria, σ factors are essential for transcription initiation. Knowledge about their regulons (i.e., genes transcribed from promoters dependent on these σ factors) is the key for understanding how bacteria cope with the changing environment and could be instrumental for biotechnologically motivated rewiring of gene expression. Here, we characterize the σI regulon from the industrially important model Gram-positive bacterium Bacillus subtilis We reveal that σI affects expression of ∼130 genes, of which 16 are directly regulated by σI, including genes encoding proteins involved in iron homeostasis. Detailed analysis of promoter elements then identifies unique sequences important for σI-dependent transcription. This study thus provides a comprehensive view on this underexplored component of the B. subtilis transcription machinery.
Keywords: RNA-seq; RNAP; iron metabolism; promoter; sigma factor.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
Identification of new sigma K-dependent promoters using an in vitro transcription system derived from Bacillus subtilis.Gene. 1999 Sep 3;237(1):45-52. doi: 10.1016/s0378-1119(99)00300-5. Gene. 1999. PMID: 10524235
-
Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches.J Mol Biol. 2002 Feb 22;316(3):443-57. doi: 10.1006/jmbi.2001.5372. J Mol Biol. 2002. PMID: 11866510
-
rpoD operon promoter used by sigma H-RNA polymerase in Bacillus subtilis.J Bacteriol. 1988 Apr;170(4):1617-21. doi: 10.1128/jb.170.4.1617-1621.1988. J Bacteriol. 1988. PMID: 3127379 Free PMC article.
-
Expression of stage II genes during sporulation in Bacillus subtilis.Res Microbiol. 1991 Sep-Oct;142(7-8):841-5. doi: 10.1016/0923-2508(91)90063-g. Res Microbiol. 1991. PMID: 1784821 Review.
-
Role of the RNA polymerase sigma subunit in transcription initiation.Res Microbiol. 2002 Nov;153(9):557-62. doi: 10.1016/s0923-2508(02)01368-2. Res Microbiol. 2002. PMID: 12455702 Review.
Cited by
-
The final proteolytic step in transmembrane signaling of multiple RsgI anti-σ factors in Clostridium thermocellum.Biosci Rep. 2025 Apr 9;45(4):233-45. doi: 10.1042/BSR20253055. Biosci Rep. 2025. PMID: 40192064 Free PMC article.
-
Essential autoproteolysis of bacterial anti-σ factor RsgI for transmembrane signal transduction.Sci Adv. 2023 Jul 7;9(27):eadg4846. doi: 10.1126/sciadv.adg4846. Epub 2023 Jul 7. Sci Adv. 2023. PMID: 37418529 Free PMC article.
-
Alternative σI/anti-σI factors represent a unique form of bacterial σ/anti-σ complex.Nucleic Acids Res. 2019 Jun 20;47(11):5988-5997. doi: 10.1093/nar/gkz355. Nucleic Acids Res. 2019. PMID: 31106374 Free PMC article.
-
Fine-Tuning Gene Expression in Bacteria by Synthetic Promoters.Methods Mol Biol. 2024;2844:179-195. doi: 10.1007/978-1-0716-4063-0_12. Methods Mol Biol. 2024. PMID: 39068340
-
Bacillus subtilis forms twisted cells with cell wall integrity defects upon removal of the molecular chaperones DnaK and trigger factor.Front Microbiol. 2023 Jan 16;13:988768. doi: 10.3389/fmicb.2022.988768. eCollection 2022. Front Microbiol. 2023. PMID: 36726573 Free PMC article.
References
-
- Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. 2001. Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci U S A 98:892–897. doi: 10.1073/pnas.98.3.892. - DOI - PMC - PubMed
-
- Rabatinová A, Šanderová H, Matějčková JJ, Korelusová J, Sojka L, Barvík I, Veronika PapouŠková Sklenár V, Žídek L, Krásný L. 2013. The δ subunit of RNA polymerase is required for rapid changes in gene expression and competitive fitness of the cell. J Bacteriol 195:2603–2611. doi: 10.1128/JB.00188-13. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

