MicroRNA-378 is involved in hedgehog-driven epithelial-to-mesenchymal transition in hepatocytes of regenerating liver
- PMID: 29915286
- PMCID: PMC6006434
- DOI: 10.1038/s41419-018-0762-z
MicroRNA-378 is involved in hedgehog-driven epithelial-to-mesenchymal transition in hepatocytes of regenerating liver
Abstract
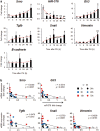
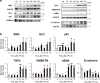
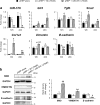
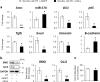
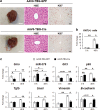
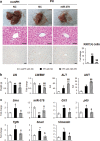
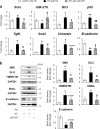
Healthy livers have a remarkable regenerative capacity for reconstructing functional hepatic parenchyma after 70% partial hepatectomy (PH). Hepatocytes, usually quiescent in normal healthy livers, proliferate to compensate for hepatic loss after PH. However, the mechanism of hepatocyte involvement in liver regeneration remains unclear. Hedgehog (Hh) pathway plays an important role in tissue reconstitution by regulating epithelial-to-mesenchymal transition (EMT) in liver disease. MicroRNA (miRNA) is involved in cell proliferation and differentiation during embryonic development and carcinogenesis. It was recently reported that miR-378 inhibits transdifferentiation of hepatic stellate cells into myofibroblasts by suppressing Gli-Krüppel family member 3 (Gli3), the Hh-target gene. We hypothesized that miR-378 influences EMT in hepatocytes by interfering with Hh signaling during liver regeneration. As hepatocytes were highly proliferative after PH in mice, miR-378 and epithelial marker, Ppar-g or E-cadherin were downregulated, whereas both Hh activators, Smoothened (Smo) and Gli3, and the EMT-inducing genes, Tgfb, Snail and Vimentin, were upregulated in the regenerating livers and in hepatocytes isolated from them. Compared to cells with or without scramble miRNA, primary hepatocytes transfected with miR-378 inhibitor contained higher levels of Gli3 with increased expression of the EMT-promoting genes, Tgfb, Snail, Col1a1, and Vimentin, suggesting that miR-378 influenced EMT in hepatocytes. Smo-depleted hepatocytes isolated from PH livers of Smo-flox mice showed downregulation of EMT-promoting genes and Gli3, with upregulation of miR-378 and E-cadherin compared to Smo-expressing hepatocytes from PH liver. In addition, delivery hepatocyte-specific AAV8 viral vector bearing Cre recombinase into Smo-flox mice impeded EMT in Smo-suppressed hepatocytes of PH liver, indicating that Smo is critical for regulating hepatocyte EMT. Furthermore, the application of miR-378 mimic into mice with PH delayed liver regeneration by interrupting hepatocyte EMT. In conclusion, our results demonstrate that miR-378 is involved in hepatocyte EMT by regulating Hh signaling during liver regeneration.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
The intrahepatic signalling niche of hedgehog is defined by primary cilia positive cells during chronic liver injury.J Hepatol. 2014 Jan;60(1):143-51. doi: 10.1016/j.jhep.2013.08.012. Epub 2013 Aug 23. J Hepatol. 2014. PMID: 23978713
-
Myofibroblastic cells function as progenitors to regenerate murine livers after partial hepatectomy.Gut. 2014 Aug;63(8):1333-44. doi: 10.1136/gutjnl-2013-305962. Epub 2013 Oct 30. Gut. 2014. PMID: 24173292 Free PMC article.
-
LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212.Cell Death Dis. 2018 Oct 3;9(10):1014. doi: 10.1038/s41419-018-1068-x. Cell Death Dis. 2018. PMID: 30282972 Free PMC article.
-
Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review).Int J Mol Med. 2008 Sep;22(3):271-5. Int J Mol Med. 2008. PMID: 18698484 Review.
-
Hepatic regeneration and the epithelial to mesenchymal transition.World J Gastroenterol. 2013 Mar 7;19(9):1380-6. doi: 10.3748/wjg.v19.i9.1380. World J Gastroenterol. 2013. PMID: 23538893 Free PMC article. Review.
Cited by
-
Mesenchymal Stem Cells Influence Activation of Hepatic Stellate Cells, and Constitute a Promising Therapy for Liver Fibrosis.Biomedicines. 2021 Nov 2;9(11):1598. doi: 10.3390/biomedicines9111598. Biomedicines. 2021. PMID: 34829827 Free PMC article. Review.
-
Tex10 promotes stemness and EMT phenotypes in esophageal squamous cell carcinoma via the Wnt/β‑catenin pathway.Oncol Rep. 2019 Dec;42(6):2600-2610. doi: 10.3892/or.2019.7376. Epub 2019 Oct 16. Oncol Rep. 2019. PMID: 31638260 Free PMC article.
-
Biomarkers for Ehlers-Danlos Syndromes: There Is a Role?Int J Mol Sci. 2021 Sep 20;22(18):10149. doi: 10.3390/ijms221810149. Int J Mol Sci. 2021. PMID: 34576312 Free PMC article. Review.
-
Customizing EV-CATCHER to Purify Placental Extracellular Vesicles from Maternal Plasma to Detect Placental Pathologies.Int J Mol Sci. 2024 May 7;25(10):5102. doi: 10.3390/ijms25105102. Int J Mol Sci. 2024. PMID: 38791142 Free PMC article.
-
HAS3-induced extracellular vesicles from melanoma cells stimulate IHH mediated c-Myc upregulation via the hedgehog signaling pathway in target cells.Cell Mol Life Sci. 2020 Oct;77(20):4093-4115. doi: 10.1007/s00018-019-03399-5. Epub 2019 Dec 9. Cell Mol Life Sci. 2020. PMID: 31820036 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

