PTPRS Regulates Colorectal Cancer RAS Pathway Activity by Inactivating Erk and Preventing Its Nuclear Translocation
- PMID: 29915291
- PMCID: PMC6006154
- DOI: 10.1038/s41598-018-27584-x
PTPRS Regulates Colorectal Cancer RAS Pathway Activity by Inactivating Erk and Preventing Its Nuclear Translocation
Abstract
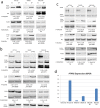
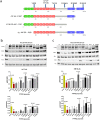
Colorectal cancer (CRC) growth and progression is frequently driven by RAS pathway activation through upstream growth factor receptor activation or through mutational activation of KRAS or BRAF. Here we describe an additional mechanism by which the RAS pathway may be modulated in CRC. PTPRS, a receptor-type protein tyrosine phosphatase, appears to regulate RAS pathway activation through ERK. PTPRS modulates ERK phosphorylation and subsequent translocation to the nucleus. Native mutations in PTPRS, present in ~10% of CRC, may reduce its phosphatase activity while increasing ERK activation and downstream transcriptional signaling.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Relationships among KRAS mutation status, expression of RAS pathway signaling molecules, and clinicopathological features and prognosis of patients with colorectal cancer.World J Gastroenterol. 2019 Feb 21;25(7):808-823. doi: 10.3748/wjg.v25.i7.808. World J Gastroenterol. 2019. PMID: 30809081 Free PMC article.
-
PDGFRA, PDGFRB, EGFR, and downstream signaling activation in malignant peripheral nerve sheath tumor.Neuro Oncol. 2009 Dec;11(6):725-36. doi: 10.1215/15228517-2009-003. Neuro Oncol. 2009. PMID: 19246520 Free PMC article.
-
ERK and AKT signaling cooperate to translationally regulate survivin expression for metastatic progression of colorectal cancer.Oncogene. 2014 Apr 3;33(14):1828-39. doi: 10.1038/onc.2013.122. Epub 2013 Apr 29. Oncogene. 2014. PMID: 23624914 Free PMC article.
-
RAS and downstream RAF-MEK and PI3K-AKT signaling in neuronal development, function and dysfunction.Biol Chem. 2016 Mar;397(3):215-22. doi: 10.1515/hsz-2015-0270. Biol Chem. 2016. PMID: 26760308 Free PMC article. Review.
-
[Roles of targeting Ras/Raf/MEK/ERK signaling pathways in the treatment of esophageal carcinoma].Yao Xue Xue Bao. 2013 May;48(5):635-41. Yao Xue Xue Bao. 2013. PMID: 23888683 Review. Chinese.
Cited by
-
Broad and thematic remodeling of the surfaceome and glycoproteome on isogenic cells transformed with driving proliferative oncogenes.Proc Natl Acad Sci U S A. 2020 Apr 7;117(14):7764-7775. doi: 10.1073/pnas.1917947117. Epub 2020 Mar 23. Proc Natl Acad Sci U S A. 2020. PMID: 32205440 Free PMC article.
-
PTPRS drives adaptive resistance to MEK/ERK inhibitors through SRC.Oncotarget. 2019 Nov 26;10(63):6768-6780. doi: 10.18632/oncotarget.27335. eCollection 2019 Nov 26. Oncotarget. 2019. PMID: 31827720 Free PMC article.
-
Erk1R84H is an oncoprotein that causes hepatocellular carcinoma in mice and imposes a rigorous negative feedback loop.Oncogene. 2025 Aug;44(31):2689-2714. doi: 10.1038/s41388-025-03437-6. Epub 2025 May 20. Oncogene. 2025. PMID: 40394416 Free PMC article.
-
Identification of New Tumor-Related Gene Mutations in Chinese Gastrointestinal Stromal Tumors.Front Cell Dev Biol. 2021 Nov 3;9:764275. doi: 10.3389/fcell.2021.764275. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34805171 Free PMC article.
-
Innovative dual system approach for selective eradication of cancer cells using viral-based delivery of natural bacterial toxin-antitoxin system.Oncogene. 2021 Aug;40(31):4967-4979. doi: 10.1038/s41388-021-01792-8. Epub 2021 Jun 25. Oncogene. 2021. PMID: 34172933 Free PMC article.
References
-
- Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2016. CA Cancer J Clin, 10.3322/caac.21332 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

