Screening and characterization of prophages in Desulfovibrio genomes
- PMID: 29915307
- PMCID: PMC6006170
- DOI: 10.1038/s41598-018-27423-z
Screening and characterization of prophages in Desulfovibrio genomes
Abstract
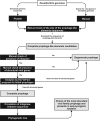
Bacteria of the genus Desulfovibrio belong to the group of Sulphate Reducing Bacteria (SRB). SRB generate significant liabilities in the petroleum industry, mainly due to their ability to microbiologically induce corrosion, biofilm formation and H2S production. Bacteriophages are an alternative control method for SRB, whose information for this group of bacteria however, is scarce. The present study developed a workflow for the identification of complete prophages in Desulfovibrio. Poly-lysogenesis was shown to be common in Desulfovibrio. In the 47 genomes analyzed 53 complete prophages were identified. These were classified within the order Caudovirales, with 69.82% belonging to the Myoviridade family. More than half the prophages identified have genes coding for lysozyme or holin. Four of the analyzed bacterial genomes present prophages with identity above 50% in the same strain, whose comparative analysis demonstrated the existence of colinearity between the sequences. Of the 17 closed bacterial genomes analyzed, 6 have the CRISPR-Cas system classified as inactive. The identification of bacterial poly-lysogeny, the proximity between the complete prophages and the possible inactivity of the CRISPR-Cas in closed bacterial genomes analyzed allowed the choice of poly-lysogenic strains with prophages belonging to the Myoviridae family for the isolation of prophages and testing of related strains for subsequent studies.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

