LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1
- PMID: 29915311
- PMCID: PMC6006356
- DOI: 10.1038/s41419-018-0759-7
LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1
Abstract
Accumulating evidence suggests long noncoding RNAs (lncRNAs) play an important role in cancer progression. However, the function of lncRNA SNHG7 in colorectal cancer (CRC) remains unclear. In this study, SNHG7 expression was significantly upregulated in CRC tissues, especially in aggressive cases. In accordance, high level of SNHG7 was observed in CRC cell lines compared to normal colon cells. Furthermore, SNHG7 overexpression promoted the proliferation, migration, and invasion of CRC cell lines, while SNHG7 depletion inhibited invasion and cell viability in vitro. Mechanistically, knockdown of SNHG7 inhibited GALNT1 and EMT markers (E-cadherin and Vimentin). Importantly, SNHG7 directly interacted with miR-216b and downregulation of miR-216b reversed efficiently the suppression of GALNT1 induced by SNHG7 siRNA. Moreover, overexpression of SNHG7 significantly enhanced the tumorigenesis and liver metastasis of SW480 cells in vivo. SNHG7 positively regulated GALNT1 level through sponging miR-216b, and played an oncogenic role in CRC progression. Together, our study elucidated the role of SNHG7 as an miRNA sponge in CRC, and shed new light on lncRNA-directed diagnostics and therapeutics in CRC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
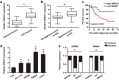

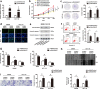
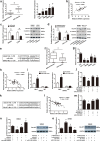


Similar articles
-
Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression.J Hematol Oncol. 2018 Jul 3;11(1):89. doi: 10.1186/s13045-018-0632-2. J Hematol Oncol. 2018. PMID: 29970122 Free PMC article.
-
LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4.Biomed Pharmacother. 2019 Oct;118:109386. doi: 10.1016/j.biopha.2019.109386. Epub 2019 Aug 30. Biomed Pharmacother. 2019. PMID: 31545291
-
Long non-coding RNA TP73-AS1 sponges miR-194 to promote colorectal cancer cell proliferation, migration and invasion via up-regulating TGFα.Cancer Biomark. 2018;23(1):145-156. doi: 10.3233/CBM-181503. Cancer Biomark. 2018. PMID: 30010111
-
SNHG7: A novel vital oncogenic lncRNA in human cancers.Biomed Pharmacother. 2020 Apr;124:109921. doi: 10.1016/j.biopha.2020.109921. Epub 2020 Jan 24. Biomed Pharmacother. 2020. PMID: 31986417 Review.
-
The role of long noncoding RNA SNHG7 in human cancers (Review).Mol Clin Oncol. 2020 Nov;13(5):45. doi: 10.3892/mco.2020.2115. Epub 2020 Aug 13. Mol Clin Oncol. 2020. PMID: 32874575 Free PMC article. Review.
Cited by
-
LncRNA SNHG6 enhances the radioresistance and promotes the growth of cervical cancer cells by sponging miR-485-3p.Cancer Cell Int. 2020 Aug 31;20:424. doi: 10.1186/s12935-020-01448-9. eCollection 2020. Cancer Cell Int. 2020. PMID: 32884447 Free PMC article.
-
Downregulation of LncRNA SNHG7 Sensitizes Colorectal Cancer Cells to Resist Anlotinib by Regulating miR-181a-5p/GATA6.Gastroenterol Res Pract. 2023 Jan 14;2023:6973723. doi: 10.1155/2023/6973723. eCollection 2023. Gastroenterol Res Pract. 2023. PMID: 36691432 Free PMC article.
-
Functional role of long non-coding RNA CASC19/miR-140-5p/CEMIP axis in colorectal cancer progression in vitro.World J Gastroenterol. 2019 Apr 14;25(14):1697-1714. doi: 10.3748/wjg.v25.i14.1697. World J Gastroenterol. 2019. PMID: 31011255 Free PMC article.
-
HOTAIR/miR-214-3p/FLOT1 axis plays an essential role in the proliferation, migration, and invasion of hepatocellular carcinoma.Int J Clin Exp Pathol. 2019 Jan 1;12(1):50-63. eCollection 2019. Int J Clin Exp Pathol. 2019. PMID: 31933720 Free PMC article.
-
Assessing the Role of Long Noncoding RNA in Nucleus Accumbens in Subjects With Alcohol Dependence.Alcohol Clin Exp Res. 2020 Dec;44(12):2468-2480. doi: 10.1111/acer.14479. Epub 2020 Nov 27. Alcohol Clin Exp Res. 2020. PMID: 33067813 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 81772277/National Natural Science Foundation of China (National Science Foundation of China)/International
- 81772277/National Natural Science Foundation of China (National Science Foundation of China)/International
- 81772277/National Natural Science Foundation of China (National Science Foundation of China)/International
- 81772277/National Natural Science Foundation of China (National Science Foundation of China)/International
- 81772277/National Natural Science Foundation of China (National Science Foundation of China)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

