Selection and Validation of Reference Genes for mRNA Expression by Quantitative Real-Time PCR Analysis in Neolamarckia cadamba
- PMID: 29915368
- PMCID: PMC6006177
- DOI: 10.1038/s41598-018-27633-5
Selection and Validation of Reference Genes for mRNA Expression by Quantitative Real-Time PCR Analysis in Neolamarckia cadamba
Abstract
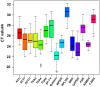
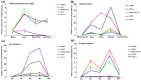
Neolamarckia cadamba is an economically-important fast-growing tree species in South China and Southeast Asia. As a prerequisite first step for future gene expression studies, we have identified and characterized a series of stable reference genes that can be used as controls for quantitative real time PCR (qRT-PCR) expression analysis in this study. The expression stability of 15 candidate reference genes in various tissues and mature leaves under different conditions was evaluated using four different algorithms, i.e., geNorm, NormFinder, BestKeeper and RefFinder. Our results showed that SAMDC was the most stable of the selected reference genes across the set of all samples, mature leaves at different photosynthetic cycles and under drought stress, whereas RPL10A had the most stable expression in various tissues. PGK and RPS25 were considered the most suitable reference for mature leaves at different developmental stages and under cold treatment, respectively. Additionally, the gene expression profiles of sucrose transporter 4 (NcSUT4), and 9-cis-epoxycarotenoid dioxygenase 3 (NcNCED3) were used to confirm the validity of candidate reference genes. Collectively, our study is the first report to validate the optimal reference genes for normalization under various conditions in N. cadamba and will benefit the future discovery of gene function in this species.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data.Sci Rep. 2019 Jun 10;9(1):8408. doi: 10.1038/s41598-019-44849-1. Sci Rep. 2019. PMID: 31182737 Free PMC article.
-
Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions.PLoS One. 2012;7(12):e51502. doi: 10.1371/journal.pone.0051502. Epub 2012 Dec 12. PLoS One. 2012. PMID: 23251557 Free PMC article.
-
Selection and validation of reference genes for quantitative real-time PCR analysis across tissues at different developmental stages in Taraxacum kok-saghyz.J Plant Physiol. 2025 Jun;309:154501. doi: 10.1016/j.jplph.2025.154501. Epub 2025 Apr 24. J Plant Physiol. 2025. PMID: 40328166
-
Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses.Gene. 2015 Jan 10;554(2):205-14. doi: 10.1016/j.gene.2014.10.049. Epub 2014 Oct 29. Gene. 2015. PMID: 25445290
-
Reference gene selection for miRNA and mRNA normalization in potato in response to potato virus Y.Mol Cell Probes. 2021 Feb;55:101691. doi: 10.1016/j.mcp.2020.101691. Epub 2020 Dec 25. Mol Cell Probes. 2021. PMID: 33358935
Cited by
-
Selection and validation of reference genes for measuring gene expression in Toona ciliata under different experimental conditions by quantitative real-time PCR analysis.BMC Plant Biol. 2020 Oct 1;20(1):450. doi: 10.1186/s12870-020-02670-3. BMC Plant Biol. 2020. PMID: 33003996 Free PMC article.
-
Population Structure and Genetic Diversity in the Natural Distribution of Neolamarckia cadamba in China.Genes (Basel). 2023 Mar 31;14(4):855. doi: 10.3390/genes14040855. Genes (Basel). 2023. PMID: 37107613 Free PMC article.
-
Selection and Validation of Appropriate Reference Genes for Quantitative RT-PCR Analysis in Rubia yunnanensis Diels Based on Transcriptome Data.Biomed Res Int. 2020 Jan 9;2020:5824841. doi: 10.1155/2020/5824841. eCollection 2020. Biomed Res Int. 2020. PMID: 31998793 Free PMC article.
-
Selection and Validation of Reference Genes for RT-qPCR in Protonemal Tissue of the Desiccation-Tolerant Moss Pseudocrossidium replicatum Under Multiple Abiotic Stress Conditions.Plants (Basel). 2025 Jun 7;14(12):1752. doi: 10.3390/plants14121752. Plants (Basel). 2025. PMID: 40573739 Free PMC article.
-
Stability evaluation and validation of appropriate reference genes for real-time PCR expression analysis of immune genes in the rohu (Labeo rohita) skin following argulosis.Sci Rep. 2023 Feb 15;13(1):2660. doi: 10.1038/s41598-023-29325-1. Sci Rep. 2023. PMID: 36792637 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

