A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors
- PMID: 29915428
- PMCID: PMC6421579
- DOI: 10.1038/s41588-018-0138-4
A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors
Abstract
We introduce and validate a new precision oncology framework for the systematic prioritization of drugs targeting mechanistic tumor dependencies in individual patients. Compounds are prioritized on the basis of their ability to invert the concerted activity of master regulator proteins that mechanistically regulate tumor cell state, as assessed from systematic drug perturbation assays. We validated the approach on a cohort of 212 gastroenteropancreatic neuroendocrine tumors (GEP-NETs), a rare malignancy originating in the pancreas and gastrointestinal tract. The analysis identified several master regulator proteins, including key regulators of neuroendocrine lineage progenitor state and immunoevasion, whose role as critical tumor dependencies was experimentally confirmed. Transcriptome analysis of GEP-NET-derived cells, perturbed with a library of 107 compounds, identified the HDAC class I inhibitor entinostat as a potent inhibitor of master regulator activity for 42% of metastatic GEP-NET patients, abrogating tumor growth in vivo. This approach may thus complement current efforts in precision oncology.
Conflict of interest statement
Competing interests
M.J.A. is Chief Scientific Officer and equity holder at DarwinHealth, Inc., a company that has licensed some of the algorithms used in this manuscript from Columbia University. A.C. is founder and equity holder of DarwinHealth Inc. Columbia University is also an equity holder in DarwinHealth Inc.
Figures
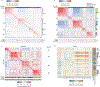




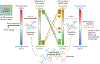
Comment in
-
H-STS, L-STS and KRJ-I are not authentic GEPNET cell lines.Nat Genet. 2019 Oct;51(10):1426-1427. doi: 10.1038/s41588-019-0490-z. Nat Genet. 2019. PMID: 31548718 No abstract available.
-
Reply to 'H-STS, L-STS and KRJ-I are not authentic GEPNET cell lines'.Nat Genet. 2019 Oct;51(10):1427-1428. doi: 10.1038/s41588-019-0509-5. Nat Genet. 2019. PMID: 31548719 Free PMC article. No abstract available.
References
-
- Weinstein IB Addiction to oncogenes–the Achilles heal of cancer. Science 297, 63–64 (2002). - PubMed
-
- Tannock IF & Hickman JA Limits to personalized cancer medicine. N. Engl. J. Med 375, 1289–1294 (2016). - PubMed
-
- Jang S & Atkins M Which drug, and when, for patients with BRAF-mutant melanoma? Lancet Oncol 14, e60–e69 (2013). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

