Diversity in the Cow Ultralong CDR H3 Antibody Repertoire
- PMID: 29915599
- PMCID: PMC5994613
- DOI: 10.3389/fimmu.2018.01262
Diversity in the Cow Ultralong CDR H3 Antibody Repertoire
Abstract
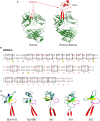
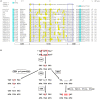
Typical antibodies found in humans and mice usually have short CDR H3s and generally flat binding surfaces. However, cows possess a subset of antibodies with ultralong CDR H3s that can range up to 70 amino acids and form a unique "stalk and knob" structure, with the knob protruding far out of the antibody surface, where it has the potential to bind antigens with concave epitopes. Activation-induced cytidine deaminase (AID) has a proven role in diversifying antibody repertoires in humoral immunity, and it has been found to induce somatic hypermutation in bovine immunoglobulin genes both before and after contact with antigen. Due to limited use of variable and diversity genes in the V(D)J recombination events that produce ultralong CDR H3 antibodies in cows, the diversity in the bovine ultralong antibody repertoire has been proposed to rely on AID-induced mutations targeted to the IGHD8-2 gene that encodes the entire knob region. In this review, we discuss the genetics, structures, and diversity of bovine ultralong antibodies, as well as the role of AID in creating a diverse antibody repertoire.
Keywords: activation-induced cytidine deaminase; antibody; antibody repertoire; cow immunoglobulin; ultralong CDRH3.
Figures
References
-
- Chehimi J, Valiante NM, D’andrea A, Rengaraju M, Rosado Z, Kobayashi M, et al. Enhancing effect of natural killer cell stimulatory factor (NKSF/interleukin-12) on cell-mediated cytotoxicity against tumor-derived and virus-infected cells. Eur J Immunol (1993) 23:1826–30. 10.1002/eji.1830230814 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

