Carbon Nanoparticles and Graphene Nanosheets as MALDI Matrices in Glycomics: a New Approach to Improve Glycan Profiling in Biological Samples
- PMID: 29916086
- PMCID: PMC6298861
- DOI: 10.1007/s13361-018-1985-z
Carbon Nanoparticles and Graphene Nanosheets as MALDI Matrices in Glycomics: a New Approach to Improve Glycan Profiling in Biological Samples
Abstract
Glycomics continues to be a highly dynamic and interesting research area due to the need to comprehensively understand the biological attributes of glycosylation in many important biological functions such as the immune response, cell development, cell differentiation/adhesion, and host-pathogen interactions. Although matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) has proven to be suitable for glycomic profiling studies, there is a need for improved sensitivity in the detection of native glycans, which ionize inefficiently. In this study, we investigated the efficiencies of graphene nanosheets (GNs) and carbon nanoparticles (CNPs) as MALDI matrices and co-matrices in glycan profiling. Our results indicated an enhancement of signal intensity by several orders of magnitude upon using GNs and CNPs in MALDI analysis of N-glycans derived from a variety of biological samples. Interestingly, increasing the amounts of CNPs and GNs improved not only the signal intensities but also prompted in-source decay (ISD) fragmentations, which produced extensive glycosidic and cross-ring cleavages. Our results indicated that the extent of ISD fragmentation could be modulated by CNP and GN concentrations, to obtain MS2 and pseudo-MS3 spectra. The results for glycan profiling in high salt solutions confirmed high salt-tolerance capacities for both CNPs and GNs. Finally, the results showed that by using CNPs and GNs as co-matrices, DHB crystal formation was more homogeneous which improved shot-to-shot reproducibility and sensitivity. Graphical Abstract ᅟ.
Keywords: Glycans; Graphene; MALDI-MS; Nanoparticles.
Figures
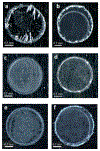

 , N-acetylglucosamine;
, N-acetylglucosamine;  , Galactose;
, Galactose;  , Fucose;
, Fucose;  , Mannose;
, Mannose;  , N-acetylneuraminic acid.
, N-acetylneuraminic acid.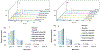

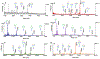
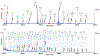
References
-
- Abou-Abbass H, Abou-El-Hassan H, Bahmad H, Zibara K, Zebian A, Youssef R, Ismail J, Zhu R, Zhou S, Dong X, Nasser M, Bahmad M, Darwish H, Mechref Y, Kobeissy F: Glycosylation and other PTMs alterations in neurodegenerative diseases: Current status and future role in neurotrauma. Electrophoresis. 37, 1549–1561 (2016) - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

