ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping
- PMID: 29916797
- PMCID: PMC6113867
- DOI: 10.1099/mgen.0.000192
ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping
Abstract
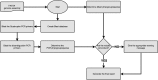
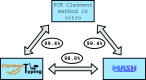
The genus Escherichia is composed of Escherichia albertii, E. fergusonii, five cryptic Escherichia clades and E. coli sensu stricto. Furthermore, the E. coli species can be divided into seven main phylogroups termed A, B1, B2, C, D, E and F. As specific lifestyles and/or hosts can be attributed to these species/phylogroups, their identification is meaningful for epidemiological studies. Classical phenotypic tests fail to identify non-sensu stricto E. coli as well as phylogroups. Clermont and colleagues have developed PCR assays that allow the identification of most of these species/phylogroups, the triplex/quadruplex PCR for E. coli phylogroup determination being the most popular. With the growing availability of whole genome sequences, we have developed the ClermonTyping method and its associated web-interface, the ClermonTyper, that allows a given strain sequence to be assigned to E. albertii, E. fergusonii, Escherichia clades I-V, E. coli sensu stricto as well as to the seven main E. coli phylogroups. The ClermonTyping is based on the concept of in vitro PCR assays and maintains the principles of ease of use and speed that prevailed during the development of the in vitro assays. This in silico approach shows 99.4 % concordance with the in vitro PCR assays and 98.8 % with the Mash genome-clustering tool. The very few discrepancies result from various errors occurring mainly from horizontal gene transfers or SNPs in the primers. We propose the ClermonTyper as a freely available resource to the scientific community at: http://clermontyping.iame-research.center/.
Keywords: E. coli; Escherichia; epidemiology; phylogeny; tool; typing.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

