YAMP: a containerized workflow enabling reproducibility in metagenomics research
- PMID: 29917068
- PMCID: PMC6047416
- DOI: 10.1093/gigascience/giy072
YAMP: a containerized workflow enabling reproducibility in metagenomics research
Abstract
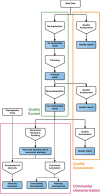
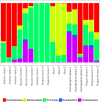
YAMP ("Yet Another Metagenomics Pipeline") is a user-friendly workflow that enables the analysis of whole shotgun metagenomic data while using containerization to ensure computational reproducibility and facilitate collaborative research. YAMP can be executed on any UNIX-like system and offers seamless support for multiple job schedulers as well as for the Amazon AWS cloud. Although YAMP was developed to be ready to use by nonexperts, bioinformaticians will appreciate its flexibility, modularization, and simple customization.
Figures


Similar articles
-
ViroProfiler: a containerized bioinformatics pipeline for viral metagenomic data analysis.Gut Microbes. 2023 Jan-Dec;15(1):2192522. doi: 10.1080/19490976.2023.2192522. Gut Microbes. 2023. PMID: 36998174 Free PMC article.
-
MGS-Fast: Metagenomic shotgun data fast annotation using microbial gene catalogs.Gigascience. 2019 Apr 1;8(4):giz020. doi: 10.1093/gigascience/giz020. Gigascience. 2019. PMID: 30942867 Free PMC article.
-
Biofilm marker discovery with cloud-based dockerized metagenomics analysis of microbial communities.Brief Bioinform. 2024 Jul 23;25(Supplement_1):bbae429. doi: 10.1093/bib/bbae429. Brief Bioinform. 2024. PMID: 39266450 Free PMC article.
-
Interrogating the microbiome: experimental and computational considerations in support of study reproducibility.Drug Discov Today. 2018 Sep;23(9):1644-1657. doi: 10.1016/j.drudis.2018.06.005. Epub 2018 Jun 8. Drug Discov Today. 2018. PMID: 29890228 Review.
-
Web Resources for Metagenomics Studies.Genomics Proteomics Bioinformatics. 2015 Oct;13(5):296-303. doi: 10.1016/j.gpb.2015.10.003. Epub 2015 Nov 18. Genomics Proteomics Bioinformatics. 2015. PMID: 26602607 Free PMC article. Review.
Cited by
-
Abnormalities in gut virome signatures linked with cognitive impairment in older adults.Gut Microbes. 2024 Jan-Dec;16(1):2431648. doi: 10.1080/19490976.2024.2431648. Epub 2024 Dec 16. Gut Microbes. 2024. PMID: 39676708 Free PMC article.
-
Red Cabbage Juice-Mediated Gut Microbiota Modulation Improves Intestinal Epithelial Homeostasis and Ameliorates Colitis.Int J Mol Sci. 2023 Dec 30;25(1):539. doi: 10.3390/ijms25010539. Int J Mol Sci. 2023. PMID: 38203712 Free PMC article.
-
Genetic and gut microbiome determinants of SCFA circulating and fecal levels, postprandial responses and links to chronic and acute inflammation.Gut Microbes. 2023 Jan-Dec;15(1):2240050. doi: 10.1080/19490976.2023.2240050. Gut Microbes. 2023. PMID: 37526398 Free PMC article.
-
Library Preparation and Sequencing Platform Introduce Bias in Metagenomic-Based Characterizations of Microbiomes.Microbiol Spectr. 2022 Apr 27;10(2):e0009022. doi: 10.1128/spectrum.00090-22. Epub 2022 Mar 15. Microbiol Spectr. 2022. PMID: 35289669 Free PMC article.
-
A metagenomic catalog for exploring the plastizymes landscape covering taxa, genes, and proteins.Sci Rep. 2023 Sep 25;13(1):16029. doi: 10.1038/s41598-023-43042-9. Sci Rep. 2023. PMID: 37749380 Free PMC article.
References
-
- Baker M. 1,500 scientists lift the lid on reproducibility. Nature News. 2016;533(7604):452. - PubMed
-
- Ioannidis JP, Allison DB, Ball CA, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41(2):149–55. - PubMed
-
- Hothorn T, Leisch F. Case studies in reproducibility. Brief Bioinform. 2011;12(3):288–300. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

