YAMP: a containerized workflow enabling reproducibility in metagenomics research
- PMID: 29917068
- PMCID: PMC6047416
- DOI: 10.1093/gigascience/giy072
YAMP: a containerized workflow enabling reproducibility in metagenomics research
Abstract
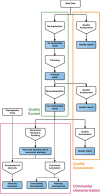
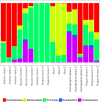
YAMP ("Yet Another Metagenomics Pipeline") is a user-friendly workflow that enables the analysis of whole shotgun metagenomic data while using containerization to ensure computational reproducibility and facilitate collaborative research. YAMP can be executed on any UNIX-like system and offers seamless support for multiple job schedulers as well as for the Amazon AWS cloud. Although YAMP was developed to be ready to use by nonexperts, bioinformaticians will appreciate its flexibility, modularization, and simple customization.
Figures


References
-
- Baker M. 1,500 scientists lift the lid on reproducibility. Nature News. 2016;533(7604):452. - PubMed
-
- Ioannidis JP, Allison DB, Ball CA, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41(2):149–55. - PubMed
-
- Hothorn T, Leisch F. Case studies in reproducibility. Brief Bioinform. 2011;12(3):288–300. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

