Perturbing cohesin dynamics drives MRE11 nuclease-dependent replication fork slowing
- PMID: 29917110
- PMCID: PMC6379725
- DOI: 10.1093/nar/gky519
Perturbing cohesin dynamics drives MRE11 nuclease-dependent replication fork slowing
Abstract
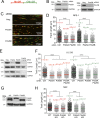
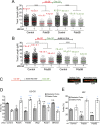
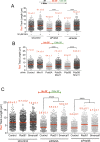
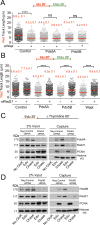
Pds5 is required for sister chromatid cohesion, and somewhat paradoxically, to remove cohesin from chromosomes. We found that Pds5 plays a critical role during DNA replication that is distinct from its previously known functions. Loss of Pds5 hinders replication fork progression in unperturbed human and mouse cells. Inhibition of MRE11 nuclease activity restores fork progression, suggesting that Pds5 protects forks from MRE11-activity. Loss of Pds5 also leads to double-strand breaks, which are again reduced by MRE11 inhibition. The replication function of Pds5 is independent of its previously reported interaction with BRCA2. Unlike Pds5, BRCA2 protects forks from nucleolytic degradation only in the presence of genotoxic stress. Moreover, our iPOND analysis shows that the loading of Pds5 and other cohesion factors on replication forks is not affected by the BRCA2 status. Pds5 role in DNA replication is shared by the other cohesin-removal factor Wapl, but not by the cohesin complex component Rad21. Interestingly, depletion of Rad21 in a Pds5-deficient background rescues the phenotype observed upon Pds5 depletion alone. These findings support a model where loss of either component of the cohesin releasin complex perturbs cohesin dynamics on replication forks, hindering fork progression and promoting MRE11-dependent fork slowing.
© The Author(s) 2018. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
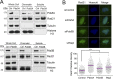
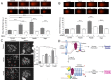
Similar articles
-
PDS5 proteins are required for proper cohesin dynamics and participate in replication fork protection.J Biol Chem. 2020 Jan 3;295(1):146-157. doi: 10.1074/jbc.RA119.011099. Epub 2019 Nov 22. J Biol Chem. 2020. PMID: 31757807 Free PMC article.
-
Brca2, Pds5 and Wapl differentially control cohesin chromosome association and function.PLoS Genet. 2018 Feb 15;14(2):e1007225. doi: 10.1371/journal.pgen.1007225. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29447171 Free PMC article.
-
The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks.Cells. 2021 Dec 8;10(12):3455. doi: 10.3390/cells10123455. Cells. 2021. PMID: 34943967 Free PMC article. Review.
-
S. cerevisiae Cells Can Grow without the Pds5 Cohesin Subunit.mBio. 2022 Aug 30;13(4):e0142022. doi: 10.1128/mbio.01420-22. Epub 2022 Jun 16. mBio. 2022. PMID: 35708277 Free PMC article.
-
The expanding phenotypes of cohesinopathies: one ring to rule them all!Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review.
Cited by
-
Mechanisms of direct replication restart at stressed replisomes.DNA Repair (Amst). 2020 Nov;95:102947. doi: 10.1016/j.dnarep.2020.102947. Epub 2020 Aug 16. DNA Repair (Amst). 2020. PMID: 32853827 Free PMC article. Review. No abstract available.
-
Human DDK rescues stalled forks and counteracts checkpoint inhibition at unfired origins to complete DNA replication.Mol Cell. 2021 Feb 4;81(3):426-441.e8. doi: 10.1016/j.molcel.2021.01.004. Mol Cell. 2021. PMID: 33545059 Free PMC article.
-
Roles for the 3D genome in the cell cycle, DNA replication, and double strand break repair.Front Cell Dev Biol. 2025 Feb 27;13:1548946. doi: 10.3389/fcell.2025.1548946. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40083661 Free PMC article. Review.
-
Pds5A and Pds5B Display Non-redundant Functions in Mitosis and Their Loss Triggers Chk1 Activation.Front Cell Dev Biol. 2020 Jul 14;8:531. doi: 10.3389/fcell.2020.00531. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32760717 Free PMC article.
-
DNA replication and replication stress response in the context of nuclear architecture.Chromosoma. 2024 Jan;133(1):57-75. doi: 10.1007/s00412-023-00813-7. Epub 2023 Dec 6. Chromosoma. 2024. PMID: 38055079 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

