N-terminal domain of human uracil DNA glycosylase (hUNG2) promotes targeting to uracil sites adjacent to ssDNA-dsDNA junctions
- PMID: 29917162
- PMCID: PMC6101581
- DOI: 10.1093/nar/gky525
N-terminal domain of human uracil DNA glycosylase (hUNG2) promotes targeting to uracil sites adjacent to ssDNA-dsDNA junctions
Abstract
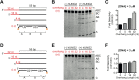
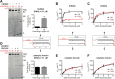
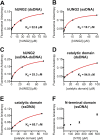
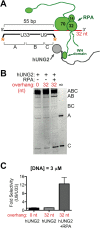
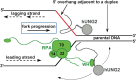
The N-terminal domain (NTD) of nuclear human uracil DNA glycosylase (hUNG2) assists in targeting hUNG2 to replication forks through specific interactions with replication protein A (RPA). Here, we explored hUNG2 activity in the presence and absence of RPA using substrates with ssDNA-dsDNA junctions that mimic structural features of the replication fork and transcriptional R-loops. We find that when RPA is tightly bound to the ssDNA overhang of junction DNA substrates, base excision by hUNG2 is strongly biased toward uracils located 21 bp or less from the ssDNA-dsDNA junction. In the absence of RPA, hUNG2 still showed an 8-fold excision bias for uracil located <10 bp from the junction, but only when the overhang had a 5' end. Biased targeting required the NTD and was not observed with the hUNG2 catalytic domain alone. Consistent with this requirement, the isolated NTD was found to bind weakly to ssDNA. These findings indicate that the NTD of hUNG2 targets the enzyme to ssDNA-dsDNA junctions using RPA-dependent and RPA-independent mechanisms. This structure-based specificity may promote efficient removal of uracils that arise from dUTP incorporation during DNA replication, or additionally, uracils that arise from DNA cytidine deamination at transcriptional R-loops during immunoglobulin class-switch recombination.
Figures

Similar articles
-
Replication Protein A Enhances Kinetics of Uracil DNA Glycosylase on ssDNA and Across DNA Junctions: Explored with a DNA Repair Complex Produced with SpyCatcher/SpyTag Ligation.Chembiochem. 2023 May 16;24(10):e202200765. doi: 10.1002/cbic.202200765. Epub 2023 Apr 18. Chembiochem. 2023. PMID: 36883884 Free PMC article.
-
Analysis of uracil DNA glycosylase (UNG2) stimulation by replication protein A (RPA) at ssDNA-dsDNA junctions.Biochim Biophys Acta Proteins Proteom. 2020 Mar;1868(3):140347. doi: 10.1016/j.bbapap.2019.140347. Epub 2019 Dec 19. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 31866506
-
hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup.J Biol Chem. 2002 Oct 18;277(42):39926-36. doi: 10.1074/jbc.M207107200. Epub 2002 Aug 2. J Biol Chem. 2002. PMID: 12161446
-
The N-terminal domain of uracil-DNA glycosylase: Roles for disordered regions.DNA Repair (Amst). 2021 May;101:103077. doi: 10.1016/j.dnarep.2021.103077. Epub 2021 Feb 18. DNA Repair (Amst). 2021. PMID: 33640758 Free PMC article. Review.
-
Chaperoning RPA during DNA metabolism.Curr Genet. 2019 Aug;65(4):857-864. doi: 10.1007/s00294-019-00945-3. Epub 2019 Feb 22. Curr Genet. 2019. PMID: 30796471 Review.
Cited by
-
Identification of a Chemotherapeutic Lead Molecule for the Potential Disruption of the FAM72A-UNG2 Interaction to Interfere with Genome Stability, Centromere Formation, and Genome Editing.Cancers (Basel). 2021 Nov 22;13(22):5870. doi: 10.3390/cancers13225870. Cancers (Basel). 2021. PMID: 34831023 Free PMC article.
-
The base excision repair process: comparison between higher and lower eukaryotes.Cell Mol Life Sci. 2021 Dec;78(24):7943-7965. doi: 10.1007/s00018-021-03990-9. Epub 2021 Nov 3. Cell Mol Life Sci. 2021. PMID: 34734296 Free PMC article. Review.
-
Ion-DNA Interactions as a Key Determinant of Uracil DNA Glycosylase Activity.Biochemistry. 2025 May 20;64(10):2332-2344. doi: 10.1021/acs.biochem.5c00067. Epub 2025 May 7. Biochemistry. 2025. PMID: 40331587 Free PMC article.
-
Facilitated Diffusion Mechanisms in DNA Base Excision Repair and Transcriptional Activation.Chem Rev. 2018 Dec 12;118(23):11298-11323. doi: 10.1021/acs.chemrev.8b00513. Epub 2018 Oct 31. Chem Rev. 2018. PMID: 30379068 Free PMC article. Review.
-
Lighting up Nobel Prize-winning studies with protein intrinsic disorder.Cell Mol Life Sci. 2022 Jul 26;79(8):449. doi: 10.1007/s00018-022-04468-y. Cell Mol Life Sci. 2022. PMID: 35882686 Free PMC article. Review.
References
-
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S.. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002; 12:1748–1755. - PubMed
-
- Nagelhus T.A., Haug T., Singh K.K., Keshav K.F., Skorpen F., Otterlei M., Bharati S., Lindmo T., Benichou S., Benarous R. et al. . A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 1997; 272:6561–6566. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

