EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration
- PMID: 29920274
- PMCID: PMC6014628
- DOI: 10.1016/j.devcel.2018.05.027
EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration
Abstract
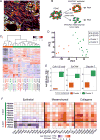
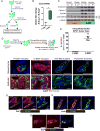
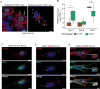
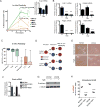
Epithelial-mesenchymal transition (EMT) is strongly implicated in tumor cell invasion and metastasis. EMT is thought to be regulated primarily at the transcriptional level through the repressive activity of EMT transcription factors. However, these classical mechanisms have been parsed out almost exclusively in vitro, leaving questions about the programs driving EMT in physiological contexts. Here, using a lineage-labeled mouse model of pancreatic ductal adenocarcinoma to study EMT in vivo, we found that most tumors lose their epithelial phenotype through an alternative program involving protein internalization rather than transcriptional repression, resulting in a "partial EMT" phenotype. Carcinoma cells utilizing this program migrate as clusters, contrasting with the single-cell migration pattern associated with traditionally defined EMT mechanisms. Moreover, many breast and colorectal cancer cell lines utilize this alternative program to undergo EMT. Collectively, these results suggest that carcinoma cells have different ways of losing their epithelial program, resulting in distinct modes of invasion and dissemination.
Keywords: E-cadherin; circulating tumor cells; collective migration; epithelial-mesenchymal transition; lineage tracing; metastasis; pancreatic cancer; partial EMT; tumor cell clusters.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interest
The authors declare no competing interests.
Figures
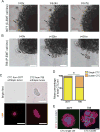
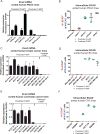
Comment in
-
EMT in Metastasis: Finding the Right Balance.Dev Cell. 2018 Jun 18;45(6):663-665. doi: 10.1016/j.devcel.2018.05.033. Dev Cell. 2018. PMID: 29920271
References
-
- Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, Younes M, Maitra A, McAllister F, Iacobuzio-Donahue CA, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2016a;35:4282–4288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

