Laser capture microdissection for transcriptomic profiles in human skin biopsies
- PMID: 29921228
- PMCID: PMC6009967
- DOI: 10.1186/s12867-018-0108-5
Laser capture microdissection for transcriptomic profiles in human skin biopsies
Abstract
Background: The acquisition of reliable tissue-specific RNA sequencing data from human skin biopsy represents a major advance in research. However, the complexity of the process of isolation of specific layers from fresh-frozen human specimen by laser capture microdissection, the abundant presence of skin nucleases and RNA instability remain relevant methodological challenges. We developed and optimized a protocol to extract RNA from layers of human skin biopsies and to provide satisfactory quality and amount of mRNA sequencing data.
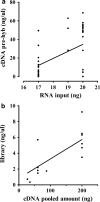
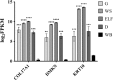
Results: The protocol includes steps of collection, embedding, freezing, histological coloration and relative optimization to preserve RNA extracted from specific components of fresh-frozen human skin biopsy of 14 subjects. Optimization of the protocol includes a preservation step in RNALater® Solution, the control of specimen temperature, the use of RNase Inhibitors and the time reduction of the staining procedure. The quality of extracted RNA was measured using the percentage of fragments longer than 200 nucleotides (DV200), a more suitable measurement for successful library preparation than the RNA Integrity Number (RIN). RNA was then enriched using the TruSeq® RNA Access Library Prep Kit (Illumina®) and sequenced on HiSeq® 2500 platform (Illumina®). Quality control on RNA sequencing data was adequate to get reliable data for downstream analysis.
Conclusions: The described implemented and optimized protocol can be used for generating transcriptomics data on skin tissues, and it is potentially applicable to other tissues. It can be extended to multicenter studies, due to the introduction of an initial step of preservation of the specimen that allowed the shipment of biological samples.
Keywords: Idiopathic neuropathy; Laser capture microdissection; RNA sequencing; Skin biopsy; Transcriptomics.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

