KDM1A microenvironment, its oncogenic potential, and therapeutic significance
- PMID: 29921310
- PMCID: PMC6006565
- DOI: 10.1186/s13072-018-0203-3
KDM1A microenvironment, its oncogenic potential, and therapeutic significance
Abstract
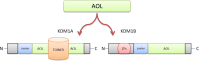
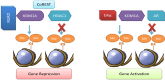
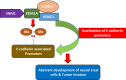
The lysine-specific histone demethylase 1A (KDM1A) was the first demethylase to challenge the concept of the irreversible nature of methylation marks. KDM1A, containing a flavin adenine dinucleotide (FAD)-dependent amine oxidase domain, demethylates histone 3 lysine 4 and histone 3 lysine 9 (H3K4me1/2 and H3K9me1/2). It has emerged as an epigenetic developmental regulator and was shown to be involved in carcinogenesis. The functional diversity of KDM1A originates from its complex structure and interactions with transcription factors, promoters, enhancers, oncoproteins, and tumor-associated genes (tumor suppressors and activators). In this review, we discuss the microenvironment of KDM1A in cancer progression that enables this protein to activate or repress target gene expression, thus making it an important epigenetic modifier that regulates the growth and differentiation potential of cells. A detailed analysis of the mechanisms underlying the interactions between KDM1A and the associated complexes will help to improve our understanding of epigenetic regulation, which may enable the discovery of more effective anticancer drugs.
Keywords: Acute myeloid leukemia; Carcinogenesis; Histone demethylation; KDM1A; TLL.
Figures

Similar articles
-
Lysine-Specific Demethylase 1A (KDM1A/LSD1): Product Recognition and Kinetic Analysis of Full-Length Histones.Biochemistry. 2016 Mar 22;55(11):1652-62. doi: 10.1021/acs.biochem.5b01135. Epub 2016 Feb 9. Biochemistry. 2016. PMID: 26673564 Free PMC article.
-
The interplay between the lysine demethylase KDM1A and DNA methyltransferases in cancer cells is cell cycle dependent.Oncotarget. 2016 Sep 13;7(37):58939-58952. doi: 10.18632/oncotarget.10624. Oncotarget. 2016. PMID: 27449289 Free PMC article.
-
A comprehensive review of lysine-specific demethylase 1 and its roles in cancer.Epigenomics. 2017 Aug;9(8):1123-1142. doi: 10.2217/epi-2017-0022. Epub 2017 Jul 12. Epigenomics. 2017. PMID: 28699367 Review.
-
Oncogenic role and target properties of the lysine-specific demethylase KDM1A in chronic lymphocytic leukemia.Blood. 2023 Jul 6;142(1):44-61. doi: 10.1182/blood.2022017230. Blood. 2023. PMID: 37023372
-
Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells.Cancer Sci. 2016 Sep;107(9):1187-92. doi: 10.1111/cas.13004. Epub 2016 Sep 1. Cancer Sci. 2016. PMID: 27375009 Free PMC article. Review.
Cited by
-
lncRNA MAGI2-AS3 Prevents the Development of HCC via Recruiting KDM1A and Promoting H3K4me2 Demethylation of the RACGAP1 Promoter.Mol Ther Nucleic Acids. 2019 Dec 6;18:351-362. doi: 10.1016/j.omtn.2019.08.020. Epub 2019 Aug 28. Mol Ther Nucleic Acids. 2019. PMID: 31629962 Free PMC article.
-
KDM1A, a potent and selective target, for the treatment of DNMT3A-deficient non-small cell lung cancer.Br J Cancer. 2024 Sep;131(4):655-667. doi: 10.1038/s41416-024-02772-x. Epub 2024 Jun 29. Br J Cancer. 2024. PMID: 38951697 Free PMC article.
-
Combining bioinformatics, cheminformatics, functional genomics and whole organism approaches for identifying epigenetic drug targets in Schistosoma mansoni.Int J Parasitol Drugs Drug Resist. 2018 Dec;8(3):559-570. doi: 10.1016/j.ijpddr.2018.10.005. Epub 2018 Nov 13. Int J Parasitol Drugs Drug Resist. 2018. PMID: 30455056 Free PMC article.
-
Molecular Characterization and Clinical Relevance of N6-Methyladenosine Regulators in Metastatic Prostate Cancer.Front Oncol. 2022 Jun 22;12:914692. doi: 10.3389/fonc.2022.914692. eCollection 2022. Front Oncol. 2022. PMID: 35814454 Free PMC article.
-
Knockdown of KDM1A suppresses tumour migration and invasion by epigenetically regulating the TIMP1/MMP9 pathway in papillary thyroid cancer.J Cell Mol Med. 2019 Aug;23(8):4933-4944. doi: 10.1111/jcmm.14311. Epub 2019 Jun 18. J Cell Mol Med. 2019. PMID: 31211500 Free PMC article.
References
-
- Perri F, Longo F, Giuliano M, Sabbatino F, Favia G, Ionna F, Addeo R, Della Vittoria Scarpati G, Di Lorenzo G, Pisconti S. Epigenetic control of gene expression: potential implications for cancer treatment. Crit Rev Oncol Hematol. 2017;111:166–172. doi: 10.1016/j.critrevonc.2017.01.020. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

