Cytokinin Signaling in Mycobacterium tuberculosis
- PMID: 29921668
- PMCID: PMC6016246
- DOI: 10.1128/mBio.00989-18
Cytokinin Signaling in Mycobacterium tuberculosis
Abstract
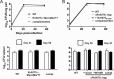
It was recently reported that the human-exclusive pathogen Mycobacterium tuberculosis secretes cytokinins, which had only been known as plant hormones. While cytokinins are well-established, adenine-based signaling molecules in plants, they have never been shown to participate in signal transduction in other kingdoms of life. M. tuberculosis is not known to interact with plants. Therefore, we tested the hypothesis that cytokinins trigger transcriptional changes within this bacterial species. Here, we show cytokinins induced the strong expression of the M. tuberculosis gene Rv0077c. We found that Rv0077c expression is repressed by a TetR-like transcriptional repressor, Rv0078. Strikingly, cytokinin-induced expression of Rv0077c resulted in a loss of acid-fast staining of M. tuberculosis While acid-fast staining is thought to be associated with changes in the bacterial cell envelope and virulence, Rv0077c-induced loss of acid-fastness did not affect antibiotic susceptibility or attenuate bacterial growth in mice, consistent with an unaltered mycolic acid profile of Rv0077c-expressing cells. Collectively, these findings show cytokinins signal transcriptional changes that can affect M. tuberculosis acid-fastness and that cytokinin signaling is no longer limited to the kingdom Plantae.IMPORTANCE Cytokinins have only previously been known as plant hormones. The discovery that they can be used as signaling molecules outside of plants broadens the repertoire of small molecules that can potentially affect gene expression in all domains of life.
Keywords: Mycobacterium tuberculosis; acid-fast staining; cytokinin; regulation; signaling.
Copyright © 2018 Samanovic et al.
Figures





References
-
- WHO 2017. Tuberculosis. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs104/en/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

