Leukemic stem cell signatures identify novel therapeutics targeting acute myeloid leukemia
- PMID: 29921955
- PMCID: PMC6889502
- DOI: 10.1038/s41408-018-0087-2
Leukemic stem cell signatures identify novel therapeutics targeting acute myeloid leukemia
Abstract
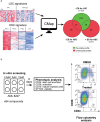
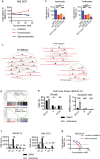
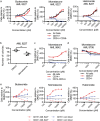
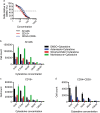
Therapy for acute myeloid leukemia (AML) involves intense cytotoxic treatment and yet approximately 70% of AML are refractory to initial therapy or eventually relapse. This is at least partially driven by the chemo-resistant nature of the leukemic stem cells (LSCs) that sustain the disease, and therefore novel anti-LSC therapies could decrease relapses and improve survival. We performed in silico analysis of highly prognostic human AML LSC gene expression signatures using existing datasets of drug-gene interactions to identify compounds predicted to target LSC gene programs. Filtering against compounds that would inhibit a hematopoietic stem cell (HSC) gene signature resulted in a list of 151 anti-LSC candidates. Using a novel in vitro LSC assay, we screened 84 candidate compounds at multiple doses and confirmed 14 drugs that effectively eliminate human AML LSCs. Three drug families presenting with multiple hits, namely antihistamines (astemizole and terfenadine), cardiac glycosides (strophanthidin, digoxin and ouabain) and glucocorticoids (budesonide, halcinonide and mometasone), were validated for their activity against human primary AML samples. Our study demonstrates the efficacy of combining computational analysis of stem cell gene expression signatures with in vitro screening to identify novel compounds that target the therapy-resistant LSC at the root of relapse in AML.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

