seq-ImmuCC: Cell-Centric View of Tissue Transcriptome Measuring Cellular Compositions of Immune Microenvironment From Mouse RNA-Seq Data
- PMID: 29922297
- PMCID: PMC5996037
- DOI: 10.3389/fimmu.2018.01286
seq-ImmuCC: Cell-Centric View of Tissue Transcriptome Measuring Cellular Compositions of Immune Microenvironment From Mouse RNA-Seq Data
Abstract
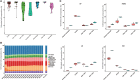
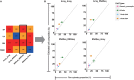
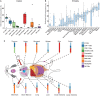
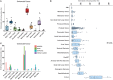
The RNA sequencing approach has been broadly used to provide gene-, pathway-, and network-centric analyses for various cell and tissue samples. However, thus far, rich cellular information carried in tissue samples has not been thoroughly characterized from RNA-Seq data. Therefore, it would expand our horizons to better understand the biological processes of the body by incorporating a cell-centric view of tissue transcriptome. Here, a computational model named seq-ImmuCC was developed to infer the relative proportions of 10 major immune cells in mouse tissues from RNA-Seq data. The performance of seq-ImmuCC was evaluated among multiple computational algorithms, transcriptional platforms, and simulated and experimental datasets. The test results showed its stable performance and superb consistency with experimental observations under different conditions. With seq-ImmuCC, we generated the comprehensive landscape of immune cell compositions in 27 normal mouse tissues and extracted the distinct signatures of immune cell proportion among various tissue types. Furthermore, we quantitatively characterized and compared 18 different types of mouse tumor tissues of distinct cell origins with their immune cell compositions, which provided a comprehensive and informative measurement for the immune microenvironment inside tumor tissues. The online server of seq-ImmuCC are freely available at http://wap-lab.org:3200/immune/.
Keywords: RNA-Seq; deconvolution; immune cell; machine learning; mouse; tumor.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

