Global transcriptome analysis to identify critical genes involved in the pathology of osteoarthritis
- PMID: 29922448
- PMCID: PMC5987685
- DOI: 10.1302/2046-3758.74.BJR-2017-0245.R1
Global transcriptome analysis to identify critical genes involved in the pathology of osteoarthritis
Abstract
Objectives: The aim of this study was to identify key pathological genes in osteoarthritis (OA).
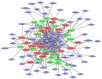
Methods: We searched and downloaded mRNA expression data from the Gene Expression Omnibus database to identify differentially expressed genes (DEGs) of joint synovial tissues from OA and normal individuals. Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analyses were used to assess the function of identified DEGs. The protein-protein interaction (PPI) network and transcriptional factors (TFs) regulatory network were used to further explore the function of identified DEGs. The quantitative real-time polymerase chain reaction (qRT-PCR) was applied to validate the result of bioinformatics analysis. Electronic validation was performed to verify the expression of selected DEGs. The diagnosis value of identified DEGs was accessed by receiver operating characteristic (ROC) analysis.
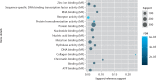
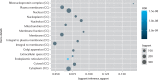
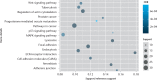
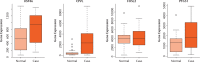
Results: A total of 1085 DEGs were identified. KEGG pathway analysis displayed that Wnt was a significantly enriched signalling pathway. Some hub genes with high interactions such as USP46, CPVL, FKBP5, FOSL2, GADD45B, PTGS1, and ZNF423 were identified in the PPI and TFs network. The results of qRT-PCR showed that GADD45B, ADAMTS1, and TFAM were down-regulated in joint synovial tissues of OA, which was consistent with the bioinformatics analysis. The expression levels of USP46, CPVL, FOSL2, and PTGS1 in electronic validation were compatible with the bio-informatics result. CPVL and TFAM had a potential diagnostic value for OA based on the ROC analysis.
Conclusion: The deregulated genes including USP46, CPVL, FKBP5, FOSL2, GADD45B, PTGS1, ZNF423, ADAMTS1, and TFAM might be involved in the pathology of OA.Cite this article: X. Zhang, Y. Bu, B. Zhu, Q. Zhao, Z. Lv, B. Li, J. Liu. Global transcriptome analysis to identify critical genes involved in the pathology of osteoarthritis. Bone Joint Res 2018;7:298-307. DOI: 10.1302/2046-3758.74.BJR-2017-0245.R1.
Keywords: Gene expression; Osteoarthritis; Protein-protein interaction network; Transcriptional factors.
Conflict of interest statement
Conflict of Interest Statement: None declared
Figures




References
-
- Kuettner KE, Cole AA. Cartilage degeneration in different human joints. Osteoarthritis Cartilage 2005;13:93-103. - PubMed
-
- Dieppe PA, Lohmander LS. Pathogenesis and management of pain in osteoarthritis. Lancet 2005;365:965-973. - PubMed
-
- Grynpas MD, Alpert B, Katz I, Lieberman I, Pritzker KP. Subchondral bone in osteoarthritis. Calcif Tissue Int 1991;49:20-26. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

