Sanguibacter massiliensis sp. nov., Actinomyces minihominis sp. nov., Clostridium minihomine sp. nov., Neobittarella massiliensis gen. nov. and Miniphocibacter massiliensis gen. nov., new bacterial species isolated by culturomics from human stool samples
- PMID: 29922471
- PMCID: PMC6004731
- DOI: 10.1016/j.nmni.2018.03.002
Sanguibacter massiliensis sp. nov., Actinomyces minihominis sp. nov., Clostridium minihomine sp. nov., Neobittarella massiliensis gen. nov. and Miniphocibacter massiliensis gen. nov., new bacterial species isolated by culturomics from human stool samples
Expression of concern in
-
Expression of Concern: Sanguibacter massiliensis sp. nov., Actinomyces minihominis sp. nov., Clostridium minihomine sp. nov., Neobittarella massiliensis gen. nov. And Miniphocibacter massiliensis gen. nov., new bacterial species isolated by culturomics from human stool samples.New Microbes New Infect. 2024 Apr 10;59:101388. doi: 10.1016/j.nmni.2024.101388. eCollection 2024 Jun. New Microbes New Infect. 2024. PMID: 38799884 Free PMC article. No abstract available.
Abstract
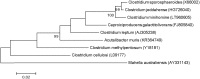
We present the description of Sanguibacter massiliensis sp. nov., Actinomyces minihominis sp. nov., Clostridium minihomine sp. nov., Neobittarella massiliensis gen. nov. and Miniphocibacter massiliensis gen. nov., new bacterial species isolated by culturomics from human stool samples.
Keywords: Actinomyces minihominis sp. nov.; Clostridium minihomine sp. nov.; Miniphocibacter massiliensis gen. nov.; Neobittarella massiliensis gen. nov.; Sanguibacter massiliensis sp. nov.
Figures




References
-
- Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–351. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
