Recent Advances in Replication and Infection of Human Parvovirus B19
- PMID: 29922597
- PMCID: PMC5996831
- DOI: 10.3389/fcimb.2018.00166
Recent Advances in Replication and Infection of Human Parvovirus B19
Abstract
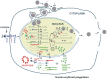
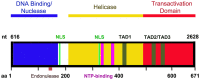
Parvovirus B19 (B19V) is pathogenic to humans and causes bone marrow failure diseases and various other inflammatory disorders. B19V infection exhibits high tropism for human erythroid progenitor cells (EPCs) in the bone marrow and fetal liver. The exclusive restriction of B19V replication to erythroid lineage cells is partly due to the expression of receptor and co-receptor(s) on the cell surface of human EPCs and partly depends on the intracellular factors essential for virus replication. We first summarize the latest developments in the viral entry process and the host cellular factors or pathways critical for B19V replication. We discuss the role of hypoxia, erythropoietin signaling and STAT5 activation in the virus replication. The B19V infection-induced DNA damage response (DDR) and cell cycle arrest at late S-phase are two key events that promote B19V replication. Lately, the virus infection causes G2 arrest, followed by the extensive cell death of EPCs that leads to anemia. We provide the current understanding of how B19V exploits the cellular resources and manipulate pathways for efficient virus replication. B19V encodes a single precursor mRNA (pre-mRNA), which undergoes alternate splicing and alternative polyadenylation to generate at least 12 different species of mRNA transcripts. The post-transcriptional processing of B19V pre-mRNA is tightly regulated through cis-acting elements and trans-acting factors flanking the splice donor or acceptor sites. Overall, in this review, we focus on the recent advances in the molecular virology and pathogenesis of B19V infection.
Keywords: DNA replication; RNA processing; erythroid precursor cells; human; infection; parvovirus B19.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

