Protein hydrogen exchange studied by the fragment separation method
- PMID: 2992314
- PMCID: PMC3442362
- DOI: 10.1016/0003-2697(85)90033-8
Protein hydrogen exchange studied by the fragment separation method
Abstract
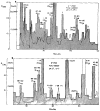
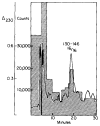
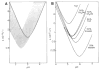
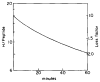
The potential of hydrogen-exchange studies for providing detailed information on protein structure and structural dynamics has not yet been realized, largely because of the continuing inability to correlate measured exchange behavior with the parts of a protein that generate that behavior. J. Rosa and F. M. Richards (1979, J. Mol. Biol. 133, 399-416) pioneered a promising approach to this problem in which tritium label at exchangeable proton sites can be located by fragmenting the protein, separating the fragments, and measuring the label carried by each fragment. However, severe losses of tritium label during the fragment separation steps have so far rendered the results ambiguous. This paper describes methods that minimize losses of tritium label during the fragment separation steps and correct for losses that do occur so that the label can be unambiguously located and even quantified. Steps that promote adequate fragment isolation are also described.
Figures



References
-
- Englander SW, Downer NW, Teitelbaum H. Annu Rev Biochem. 1972;41:903–924. - PubMed
-
- Englander SW, Kallenbach NR. Quart Rev Biophys. 1984;16:521–655. - PubMed
-
- Woodward CK, Simon T, Tuchsen E. Mol Cell Biochem. 1982;48:135–160. - PubMed
-
- Kim PS, Baldwin RL. Annu Rev Biochem. 1982;51:459–489. - PubMed
-
- Allewell NM. J Biochem Biophys Methods. 1983;7:345–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

