Dynamic Nuclear Polarization Nuclear Magnetic Resonance in Human Cells Using Fluorescent Polarizing Agents
- PMID: 29924582
- PMCID: PMC6842659
- DOI: 10.1021/acs.biochem.8b00257
Dynamic Nuclear Polarization Nuclear Magnetic Resonance in Human Cells Using Fluorescent Polarizing Agents
Abstract
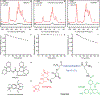
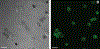
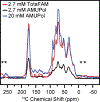
Solid state nuclear magnetic resonance (NMR) enables atomic-resolution characterization of the molecular structure and dynamics within complex heterogeneous samples, but it is typically insensitive. Dynamic nuclear polarization (DNP) increases the NMR signal intensity by orders of magnitude and can be performed in combination with magic angle spinning (MAS) for sensitive, high-resolution spectroscopy. Here we report MAS DNP experiments, for the first time, within intact human cells with >40-fold DNP enhancement and a sample temperature of <6 K. In addition to cryogenic MAS results at <6 K, we also show in-cell DNP enhancements of 57-fold at 90 K. In-cell DNP is demonstrated using biradicals and sterically shielded monoradicals as polarizing agents. A novel trimodal polarizing agent is introduced for DNP, which contains a nitroxide biradical, a targeting peptide for cell penetration, and a fluorophore for subcellular localization with confocal microscopy. The fluorescent polarizing agent provides in-cell DNP enhancements of 63-fold at a concentration of 2.7 mM. These experiments pave the way for structural characterization of biomolecules in an endogenous cellular context.
Figures


References
-
- Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VMY, George JM, and Rienstra CM (2016) Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol 23, 1–9. - PMC - PubMed
-
- Zech SG, Wand AJ, and McDermott AE (2005) Protein Structure Determination by High-Resolution Solid-State NMR Spectroscopy: Application to Microcrystalline Ubiquitin. J. Am. Chem. Soc 127, 8618–8626. - PubMed
-
- Thongsomboon W, Serra DO, Possling A, Hadjineophytou C, Hengge R, and Cegelski L (2018) Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science 359, 334–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

