Hansenula polymorpha Aat2p is targeted to peroxisomes via a novel Pex20p-dependent pathway
- PMID: 29924881
- PMCID: PMC6099311
- DOI: 10.1002/1873-3468.13168
Hansenula polymorpha Aat2p is targeted to peroxisomes via a novel Pex20p-dependent pathway
Abstract
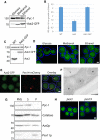
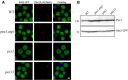
Saccharomyces cerevisiae Aat2p contains a peroxisomal targeting signal type-1 and localizes to peroxisomes in oleate-grown cells, but not in glucose-grown cells. Here, we have investigated Aat2p from the yeast Hansenula polymorpha, which lacks a recognizable peroxisomal targeting signal. Aat2p tagged with GFP at its C terminus displays a dual cytosol-peroxisome localization in ethanol-grown cells. The partial peroxisomal localization of Aat2p persisted in the absence of the classical cycling receptors Pex5p and Pex7p but Aat2p targeting to peroxisomes was reduced in cells deleted for the matrix protein import factors PEX1, PEX2 and PEX13. Furthermore, we demonstrate that Aat2p targeting to peroxisomes requires Pex20p. Together, our data identify a Pex20p-dependent pathway for targeting Aat2p to peroxisomes.
Keywords: Hansenula polymorpha; Aat2p; Peroxisome protein import; Pex20p; aspartate aminotransferase.
© 2018 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures


References
-
- Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB and Moser HW (2006) Peroxisome biogenesis disorders. Biochim Biophys Acta 1763, 1733–1748. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

