Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity
- PMID: 29924976
- PMCID: PMC6344351
- DOI: 10.1016/j.immuni.2018.05.013
Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity
Abstract
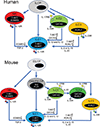
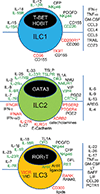
Type 1, 2, and 3 innate lymphoid cells (ILCs) have emerged as tissue-resident innate correlates of T helper 1 (Th1), Th2, and Th17 cells. Recent studies suggest that ILCs are more diverse than originally proposed; this might reflect truly distinct lineages or adaptation of ILCs to disparate tissue microenvironments, known as plasticity. Given that ILCs strikingly resemble T cells, are they redundant? While the regulation, timing, and magnitude of ILC and primary T cell responses differ, tissue-resident memory T cells may render ILCs redundant during secondary responses. The unique impact of ILCs in immunity is probably embodied in the extensive array of surface and intracellular receptors that endow these cells with the ability to distinguish between normal and pathogenic components, interact with other cells, and calibrate their cytokine secretion accordingly. Here I review recent advances in elucidating the diversity of ILCs and discuss their unique and redundant functions.
Copyright © 2018. Published by Elsevier Inc.
Figures

References
-
- Bank U, Deiser K, Finke D, Hammerling GJ, Arnold B, Schuler T. Cutting edge: innate lymphoid cells suppress homeostatic T cell expansion in neonatal mice. J Immunol. 2016;196:3532–3536. - PubMed
-
- Bar-Ephraim YE, Mebius RE. Innate lymphoid cells in secondary lymphoid organs. Immunol Rev. 2016;271:185–199. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

