Thermodynamics of Conformational Transitions in a Disordered Protein Backbone Model
- PMID: 29925017
- PMCID: PMC6026333
- DOI: 10.1016/j.bpj.2018.04.027
Thermodynamics of Conformational Transitions in a Disordered Protein Backbone Model
Abstract
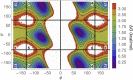
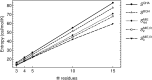
Conformational entropy is expected to contribute significantly to the thermodynamics of structural transitions in intrinsically disordered proteins or regions in response to protein/ligand binding, posttranslational modifications, and environmental changes. We calculated the backbone (dihedral) conformational entropy of oligoglycine (GlyN), a protein backbone mimic and model intrinsically disordered region, as a function of chain length (N=3, 4, 5, 10, and 15) from simulations using three different approaches. The backbone conformational entropy scales linearly with chain length with a slope consistent with the entropy of folding of well-structured proteins. The entropic contributions of second-order dihedral correlations are predominantly through intraresidue ϕ-ψ pairs, suggesting that oligoglycine may be thermodynamically modeled as a system of independent glycine residues. We find the backbone conformational entropy to be largely independent of global structural parameters, like the end-to-end distance and radius of gyration. We introduce a framework referred to herein as "ensemble confinement" to estimate the loss (gain) of conformational free energy and its entropic component when individual residues are constrained to (released from) particular regions of the ϕ-ψ map. Quantitatively, we show that our protein backbone model resists ordering/folding with a significant, unfavorable ensemble confinement free energy because of the loss of a substantial portion of the absolute backbone entropy. Proteins can couple this free-energy reservoir to distal binding events as a regulatory mechanism to promote or suppress binding.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Comment in
-
Tuning Free Energy by Backbone Conformational Entropy: A Strategy from Disordered Proteins.Biophys J. 2018 Jun 19;114(12):2757-2758. doi: 10.1016/j.bpj.2018.05.019. Biophys J. 2018. PMID: 29925011 Free PMC article. No abstract available.
References
-
- Dunker A.K., Brown C.J., Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. - PubMed
-
- Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. - PubMed
-
- Uversky V.N., Oldfield C.J., Dunker A.K. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18:343–384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

