Detailed genetic characteristics of an international large cohort of patients with Stargardt disease: ProgStar study report 8
- PMID: 29925512
- PMCID: PMC6579578
- DOI: 10.1136/bjophthalmol-2018-312064
Detailed genetic characteristics of an international large cohort of patients with Stargardt disease: ProgStar study report 8
Abstract
Background/aims: To describe the genetic characteristics of the cohort enrolled in the international multicentre progression of Stargardt disease 1 (STGD1) studies (ProgStar) and to determine geographic differences based on the allele frequency.
Methods: 345 participants with a clinical diagnosis of STGD1 and harbouring at least one disease-causing ABCA4 variant were enrolled from 9 centres in the USA and Europe. All variants were reviewed and in silico analysis was performed including allele frequency in public databases and pathogenicity predictions. Participants with multiple likely pathogenic variants were classified into four national subgroups (USA, UK, France, Germany), with subsequent comparison analysis of the allele frequency for each prevalent allele.
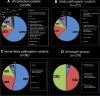
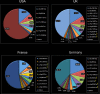
Results: 211 likely pathogenic variants were identified in the total cohort, including missense (63%), splice site alteration (18%), stop (9%) and others. 50 variants were novel. Exclusively missense variants were detected in 139 (50%) of 279 patients with multiple pathogenic variants. The three most prevalent variants of these patients with multiple pathogenic variants were p.G1961E (15%), p.G863A (7%) and c.5461-10 T>C (5%). Subgroup analysis revealed a statistically significant difference between the four recruiting nations in the allele frequency of nine variants.
Conclusions: There is a large spectrum of ABCA4 sequence variants, including 50 novel variants, in a well-characterised cohort thereby further adding to the unique allelic heterogeneity in STGD1. Approximately half of the cohort harbours missense variants only, indicating a relatively mild phenotype of the ProgStar cohort. There are significant differences in allele frequencies between nations, although the three most prevalent variants are shared as frequent variants.
Keywords: genetics; macula; retina.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: KF is a a paid consultant of Astellas Pharma, Kubota Pharmaceutical Holdings. DGB is a consultant for NightStaRX, AGTC, Shire, Ionis and Genentech. EZ is a member of the Data Monitoring and Safety Board/Committee of the following entities: ReNeuron Group/Ora, NightStaRx, RD-CURE Consortium and principal investigator in clinical trials sponsored by QLT, SHIRE and FFB at the University of Tuebingen, Institute for Ophthalmic Research. HPNS is a paid consultant of the following entities: Boehringer Ingelheim Pharma, Daiichi Sankyo, Gerson Lehrman Group; Guidepoint and Shire. HPNS is member of the Scientific Advisory Board of the Astellas Institute for Regenerative Medicine; Gensight Biologics; Vision Medicines and Intellia Therapeutics. HPNS is member of the Data Monitoring and Safety Board/Committee of the following entities: Genentech/F. Hoffmann-La Roche and ReNeuron Group/Ora. These arrangements have been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies. Johns Hopkins University and Bayer Pharma have an active research collaboration and option agreement. These arrangements have also been reviewed and approved by the University of Basel (Universitätsspital Basel, USB) in accordance with its conflict of interest policies. HPNS is principal investigator of grants at the USB sponsored by the following entity: Acucela; NightstaRx; QLT. Grants at USB are negotiated and administered by the institution (USB) which receives them on its proper accounts.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
