Fission Yeast CENP-C (Cnp3) Plays a Role in Restricting the Site of CENP-A Accumulation
- PMID: 29925533
- PMCID: PMC6071599
- DOI: 10.1534/g3.118.200486
Fission Yeast CENP-C (Cnp3) Plays a Role in Restricting the Site of CENP-A Accumulation
Abstract
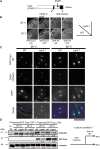
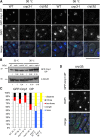
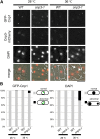
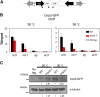
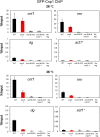
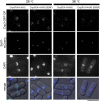
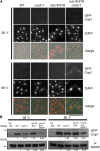
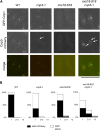
The centromere is a chromosomal locus where a microtubule attachment site, termed kinetochore, is assembled in mitosis. In most eukaryotes, with the exception of holocentric species, each chromosome contains a single distinct centromere. A chromosome with an additional centromere undergoes successive rounds of anaphase bridge formation and breakage, or triggers a cell cycle arrest imposed by DNA damage and replication checkpoints. We report here a study in Schizosaccharomyces pombe to characterize a mutant (cnp3-1) in a gene encoding a homolog of mammalian centromere-specific protein, CENP-C. At the restrictive temperature 36°, the Cnp3-1 mutant protein loses its localization at the centromere. In the cnp3-1 mutant, the level of the Cnp1 (a homolog of a centromere-specific histone CENP-A) also decreases at the centromere. Interestingly, the cnp3-1 mutant is prone to promiscuous accumulation of Cnp1 at non-centromeric regions, when Cnp1 is present in excess. Unlike the wild type protein, Cnp3-1 mutant protein is found at the sites of promiscuous accumulation of Cnp1, suggesting that Cnp3-1 may stabilize or promote accumulation of Cnp1 at non-centromeric regions. From these results, we infer the role of Cnp3 in restricting the site of accumulation of Cnp1 and thus to prevent formation of de novo centromeres.
Keywords: CENP-A; CENP-C; and fission yeast; centromere.
Copyright © 2018 Suma et al.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
