EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 Act in a Regulatory Loop That Synergistically Modulates Ethylene Biosynthesis and Anthocyanin Accumulation
- PMID: 29925585
- PMCID: PMC6181056
- DOI: 10.1104/pp.18.00068
EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 Act in a Regulatory Loop That Synergistically Modulates Ethylene Biosynthesis and Anthocyanin Accumulation
Abstract
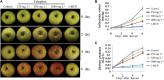
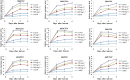
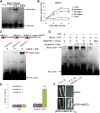
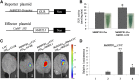
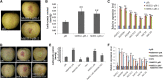
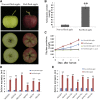
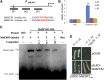
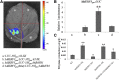
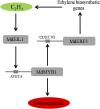
Ethylene regulates climacteric fruit ripening, and EIN3-LIKE1 (EIL1) plays an important role in this process. In apple (Malus domestica), fruit coloration is accompanied by ethylene release during fruit ripening, but the molecular mechanism that underlies these two physiological processes is unknown. In this study, we found that ethylene treatment markedly induced fruit coloration as well as the expression of MdMYB1, a positive regulator of anthocyanin biosynthesis and fruit coloration. In addition, we found that MdEIL1 directly bound to the promoter of MdMYB1 and transcriptionally activated its expression, which resulted in anthocyanin biosynthesis and fruit coloration. Furthermore, MdMYB1 interacted with the promoter of ETHYLENE RESPONSE FACTOR3, a key regulator of ethylene biosynthesis, thereby providing a positive feedback for ethylene biosynthesis regulation. Overall, our findings provide insight into a mechanism involving the synergistic interaction of the ethylene signal with the MdMYB1 transcription factor to regulate ethylene biosynthesis and fruit coloration in apple.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures
References
-
- Adams-Phillips L, Barry C, Giovannoni J (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci 9: 331–338 - PubMed
-
- Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 - PubMed
-
- Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13: 99–102 - PubMed
-
- Alonso JM, Stepanova AN (2004) The ethylene signaling pathway. Science 306: 1513–1515 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

