Droplet Digital PCR-Based Detection of Clarithromycin Resistance in Helicobacter pylori Isolates Reveals Frequent Heteroresistance
- PMID: 29925646
- PMCID: PMC6113488
- DOI: 10.1128/JCM.00019-18
Droplet Digital PCR-Based Detection of Clarithromycin Resistance in Helicobacter pylori Isolates Reveals Frequent Heteroresistance
Abstract
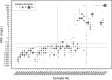
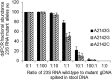
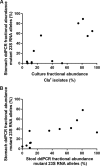
Chronic infection with Helicobacter pylori causes peptic ulcers and stomach cancer in a subset of infected individuals. While standard eradication therapy includes multiple antibiotics, treatment failure due to resistance is an increasing clinical problem. Accurate assessment of H. pylori antimicrobial resistance has been limited by slow growth and sampling of few isolates per subject. We established a method to simultaneously quantify H. pylori clarithromycin-resistant (mutant) and -susceptible (wild-type) 23S rRNA gene alleles in both stomach and stool samples using droplet digital PCR (ddPCR). In 49 subjects, we assessed the performance of these assays alongside clarithromycin MIC testing of up to 16 H. pylori isolates per subject and included both cancer (25 subjects) and noncancer (24 subjects) cases. Gastric ddPCR and H. pylori culture showed agreement with urea breath test (UBT) detection of infection in 94% and 88% of subjects, respectively, while stool ddPCR showed agreement with UBT in 92% of subjects. Based on MIC testing of 43 culture-positive cases, 20 subjects had only susceptible isolates, 14 had a mix of susceptible and resistant isolates, and 9 had only resistant isolates. ddPCR of gastric samples indicated that 21 subjects had only wild-type alleles, 13 had a mixed genotype, and 9 had only mutant alleles. Stool ddPCR detected mutant alleles in four subjects for which mutant alleles were not detected by stomach ddPCR, and no resistant isolates were cultured. Our results indicate that ddPCR detects H. pylori clarithromycin resistance-associated genotypes, especially in the context of heteroresistance.
Keywords: Helicobacter pylori; clarithromycin resistance; droplet digital PCR; heteroresistance; noninvasive detection.
Copyright © 2018 American Society for Microbiology.
Figures
References
-
- Nishizawa T, Maekawa T, Watanabe N, Harada N, Hosoda Y, Yoshinaga M, Yoshio T, Ohta H, Inoue S, Toyokawa T, Yamashita H, Saito H, Kuwai T, Katayama S, Masuda E, Miyabayashi H, Kimura T, Nishizawa Y, Takahashi M, Suzuki H. 2015. Clarithromycin versus metronidazole as first-line Helicobacter pylori eradication: a multicenter, prospective, randomized controlled study in Japan. J Clin Gastroenterol 49:468–471. doi:10.1097/MCG.0000000000000165. - DOI - PubMed
-
- Aguilera-Correa JJ, Urruzuno P, Barrio J, Martinez MJ, Agudo S, Somodevilla A, Llorca L, Alarcón T. 2017. Detection of Helicobacter pylori and the genotypes of resistance to clarithromycin and the heterogeneous genotype to this antibiotic in biopsies obtained from symptomatic children. Diagn Microbiol Infect Dis 87:150–153. doi:10.1016/j.diagmicrobio.2016.03.001. - DOI - PubMed
-
- Fontana C, Favaro M, Minelli S, Criscuolo AA, Pietroiusti A, Galante A, Favalli C. 2002. New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob Agents Chemother 46:3765–3769. doi:10.1128/AAC.46.12.3765-3769.2002. - DOI - PMC - PubMed
-
- Kim JM, Kim JS, Kim N, Kim YJ, Kim IY, Chee YJ, Lee CH, Jung HC. 2008. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J Microbiol Biotechnol 18:1584–1589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

