Pigs Lacking the Scavenger Receptor Cysteine-Rich Domain 5 of CD163 Are Resistant to Porcine Reproductive and Respiratory Syndrome Virus 1 Infection
- PMID: 29925651
- PMCID: PMC6069206
- DOI: 10.1128/JVI.00415-18
Pigs Lacking the Scavenger Receptor Cysteine-Rich Domain 5 of CD163 Are Resistant to Porcine Reproductive and Respiratory Syndrome Virus 1 Infection
Erratum in
-
Erratum for Burkard et al., "Pigs Lacking the Scavenger Receptor Cysteine-Rich Domain 5 of CD163 Are Resistant to Porcine Reproductive and Respiratory Syndrome Virus 1 Infection".J Virol. 2020 Jul 16;94(15):e00951-20. doi: 10.1128/JVI.00951-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32675378 Free PMC article. No abstract available.
Abstract
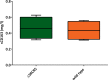
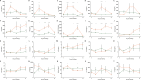
Porcine reproductive and respiratory syndrome virus (PRRSV) has a narrow host cell tropism, limited to cells of the monocyte/macrophage lineage. CD163 protein is expressed at high levels on the surface of specific macrophage types, and a soluble form is circulating in blood. CD163 has been described as a fusion receptor for PRRSV, with the scavenger receptor cysteine-rich domain 5 (SRCR5) region having been shown to be the interaction site for the virus. As reported previously, we have generated pigs in which exon 7 of the CD163 gene has been deleted using CRISPR/Cas9 editing in pig zygotes. These pigs express CD163 protein lacking SRCR5 (ΔSRCR5 CD163) and show no adverse effects when maintained under standard husbandry conditions. Not only was ΔSRCR5 CD163 detected on the surface of macrophage subsets, but the secreted, soluble protein can also be detected in the serum of the edited pigs, as shown here by a porcine soluble CD163-specific enzyme-linked immunosorbent assay (ELISA). Previous results showed that primary macrophage cells from ΔSRCR5 CD163 animals are resistant to PRRSV-1 subtype 1, 2, and 3 as well as PRRSV-2 infection in vitro Here, ΔSRCR5 pigs were challenged with a highly virulent PRRSV-1 subtype 2 strain. In contrast to the wild-type control group, ΔSRCR5 pigs showed no signs of infection and no viremia or antibody response indicative of a productive infection. Histopathological analysis of lung and lymph node tissue showed no presence of virus-replicating cells in either tissue. This shows that ΔSRCR5 pigs are fully resistant to infection by the virus.IMPORTANCE Porcine reproductive and respiratory syndrome (PRRS) virus (PRRSV) is the etiological agent of PRRS, causing late-term abortions, stillbirths, and respiratory disease in pigs, incurring major economic losses to the worldwide pig industry. The virus is highly mutagenic and can be divided into two species, PRRSV-1 and PRRSV-2, each containing several subtypes. Current control strategies mainly involve biosecurity measures, depopulation, and vaccination. Vaccines are at best only partially protective against infection with heterologous subtypes and sublineages, and modified live vaccines have frequently been reported to revert to virulence. Here, we demonstrate that a genetic-control approach results in complete resistance to PRRSV infection in vivo CD163 is edited so as to remove the viral interaction domain while maintaining protein expression and biological function, averting any potential adverse effect associated with protein knockout. This research demonstrates a genetic-control approach with potential benefits in animal welfare as well as to the pork industry.
Keywords: CD163; CRISPR/Cas9; PRRSV; arterivirus; exon deletion; genome editing; nidovirus; porcine reproductive and respiratory syndrome virus; resistance.
Copyright © 2018 Burkard et al.
Figures


References
-
- Christianson WT, Joo HS. 1994. Porcine reproductive and respiratory syndrome: a review. J Swine Health Prod 2:10–28.
-
- Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK, Wang C, Yeske PE, Mowrer CL, Hayley CA. 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Health Prod 21:72–84.
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/D/05251442/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05251443/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20002172/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L004143/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20002174/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

